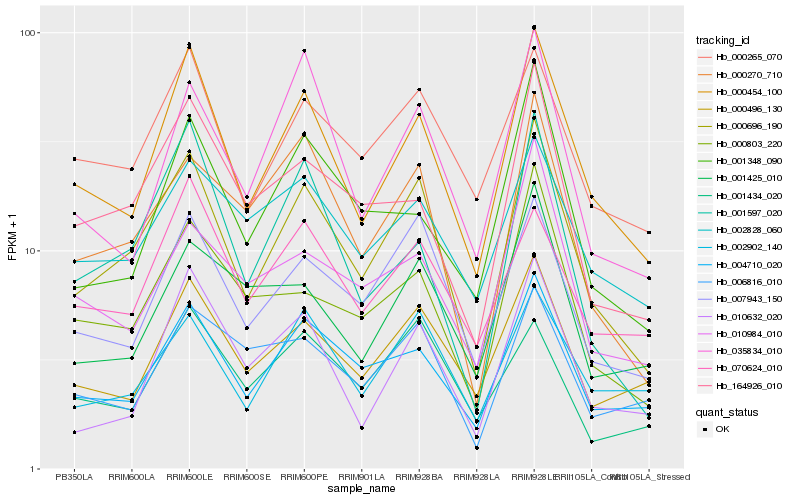
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000696_190 |
0.0 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 1 [Jatropha curcas] |
| 2 |
Hb_000454_100 |
0.0972489086 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 3 |
Hb_007943_150 |
0.1119602535 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_004710_020 |
0.1173991707 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 5 |
Hb_000270_710 |
0.1179993545 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000265_070 |
0.1285879968 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 7 |
Hb_002902_140 |
0.134490173 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 8 |
Hb_000496_130 |
0.1353859793 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 9 |
Hb_000803_220 |
0.1364872799 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlD, chloroplastic [Jatropha curcas] |
| 10 |
Hb_164926_010 |
0.1381052363 |
- |
- |
Granule-bound starch synthase 1, chloroplastic/amyloplastic [Gossypium arboreum] |
| 11 |
Hb_001597_020 |
0.142576298 |
- |
- |
PREDICTED: uncharacterized protein LOC105629846 isoform X2 [Jatropha curcas] |
| 12 |
Hb_035834_010 |
0.1445614106 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_002828_060 |
0.1476284335 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_010632_020 |
0.1484124344 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 15 |
Hb_001348_090 |
0.1531357337 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001425_010 |
0.1547880493 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 17 |
Hb_070624_010 |
0.1548609394 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 18 |
Hb_010984_010 |
0.1549978322 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_006816_010 |
0.1561014042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001434_020 |
0.1563112687 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |