| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
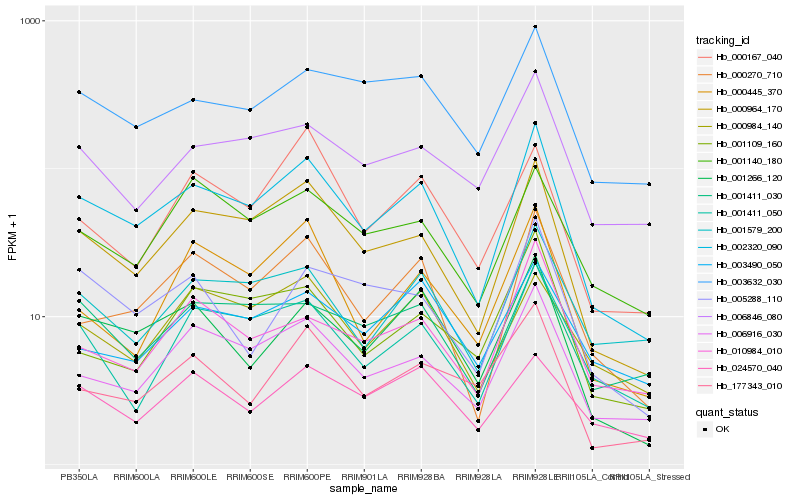
Hb_002320_090 |
0.0 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 2 |
Hb_000964_170 |
0.0743867542 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 3 |
Hb_006916_030 |
0.104485026 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 4 |
Hb_000270_710 |
0.1057198066 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001266_120 |
0.1135545215 |
- |
- |
auxilin, putative [Ricinus communis] |
| 6 |
Hb_003632_030 |
0.1290886802 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 7 |
Hb_001411_050 |
0.1294737001 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 8 |
Hb_001579_200 |
0.1321984837 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 9 |
Hb_001140_180 |
0.1358134043 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 10 |
Hb_006846_080 |
0.1358737397 |
- |
- |
calnexin, putative [Ricinus communis] |
| 11 |
Hb_001411_030 |
0.1373794912 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 12 |
Hb_010984_010 |
0.1399289247 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000984_140 |
0.141339684 |
- |
- |
PREDICTED: xylulose kinase [Jatropha curcas] |
| 14 |
Hb_005288_110 |
0.1422326249 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 15 |
Hb_000445_370 |
0.1422897312 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 16 |
Hb_001109_160 |
0.1445936557 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_177343_010 |
0.1451622898 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 18 |
Hb_000167_040 |
0.1475130146 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 19 |
Hb_024570_040 |
0.1512529522 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 20 |
Hb_003490_050 |
0.1520021725 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |