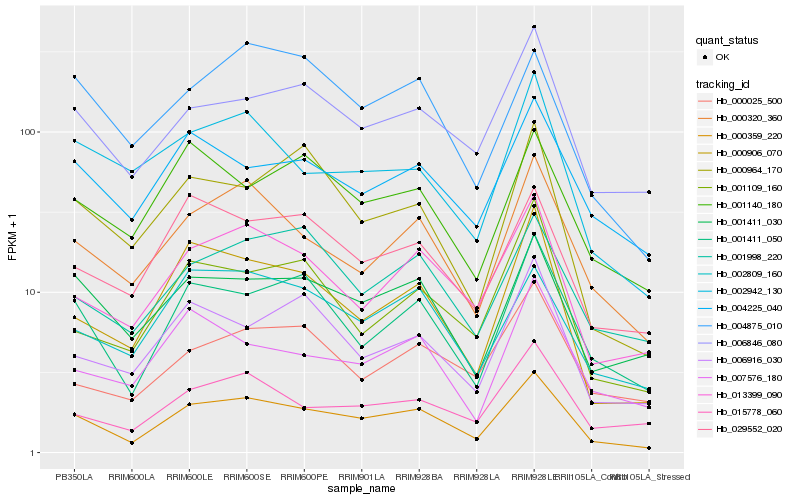
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_220 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000320_360 |
0.1070487238 |
- |
- |
PREDICTED: starch synthase 3, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_001411_050 |
0.107245811 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 4 |
Hb_013399_090 |
0.1122054192 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 5 |
Hb_000906_070 |
0.116032722 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_006846_080 |
0.117404446 |
- |
- |
calnexin, putative [Ricinus communis] |
| 7 |
Hb_001411_030 |
0.1182119185 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |
| 8 |
Hb_002809_160 |
0.1301970538 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 9 |
Hb_029552_020 |
0.130678927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004225_040 |
0.1338815808 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Populus euphratica] |
| 11 |
Hb_015778_060 |
0.1352532833 |
- |
- |
PREDICTED: uncharacterized protein LOC105650696 [Jatropha curcas] |
| 12 |
Hb_002942_130 |
0.1360965458 |
- |
- |
PREDICTED: chaperone protein ClpC, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001998_220 |
0.1372367355 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000964_170 |
0.1379861266 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 15 |
Hb_004875_010 |
0.1384357128 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 16 |
Hb_006916_030 |
0.1387516618 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 17 |
Hb_001109_160 |
0.139583304 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001140_180 |
0.1431493094 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 19 |
Hb_007576_180 |
0.1445693896 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 20 |
Hb_000025_500 |
0.1462982666 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |