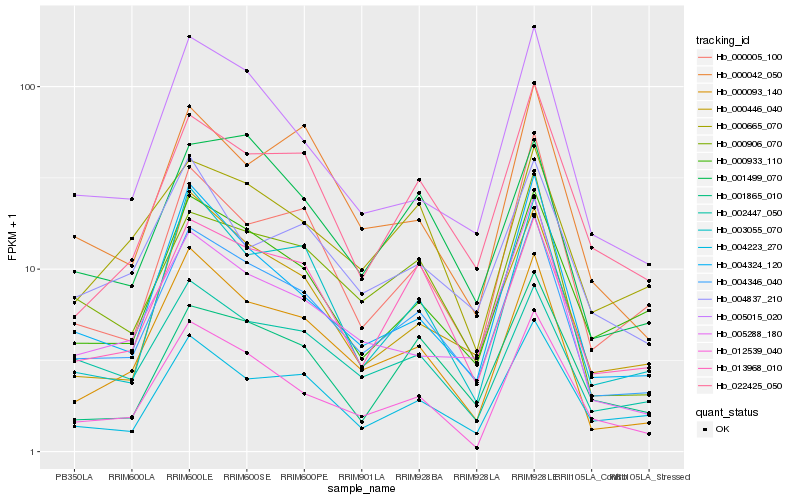
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004346_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 2 |
Hb_000093_140 |
0.0802061267 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000906_070 |
0.0947410544 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005015_020 |
0.0978094052 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_013968_010 |
0.1001400881 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 6 |
Hb_004324_120 |
0.1009860355 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 7 |
Hb_002447_050 |
0.1043307602 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_012539_040 |
0.1044404405 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 9 |
Hb_000933_110 |
0.1051483436 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 10 |
Hb_000446_040 |
0.1078369805 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 11 |
Hb_005288_180 |
0.1093486954 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_022425_050 |
0.1097233001 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 13 |
Hb_004223_270 |
0.1117694263 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000005_100 |
0.1142773106 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001499_070 |
0.1161437077 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 16 |
Hb_003055_070 |
0.1187905822 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_004837_210 |
0.1207778773 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 18 |
Hb_000042_050 |
0.1234095278 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001865_010 |
0.1236596427 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 20 |
Hb_000665_070 |
0.1238743502 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |