| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
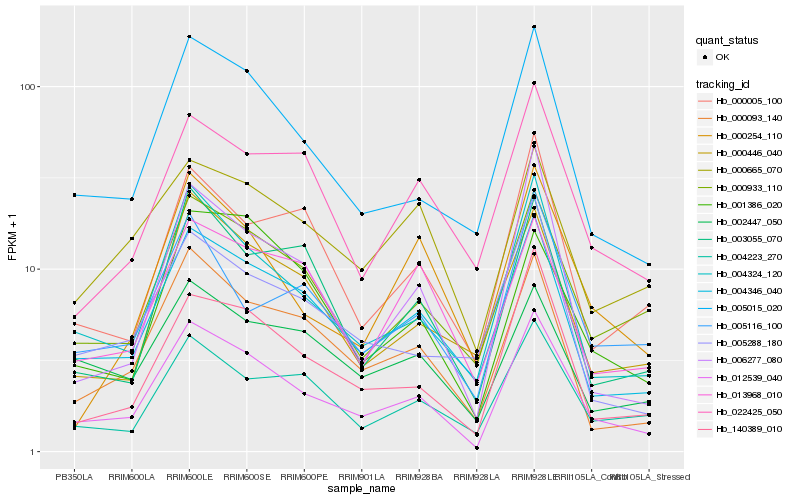
Hb_012539_040 |
0.0 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 2 |
Hb_004324_120 |
0.0959642463 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 3 |
Hb_005015_020 |
0.1039833313 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_004346_040 |
0.1044404405 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 5 |
Hb_140389_010 |
0.1160844977 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_000093_140 |
0.1198324709 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000933_110 |
0.1258665917 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 8 |
Hb_004223_270 |
0.13856853 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006277_080 |
0.1393529607 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 10 |
Hb_000446_040 |
0.1413280981 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 11 |
Hb_003055_070 |
0.1462251998 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_000005_100 |
0.1470428651 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001386_020 |
0.1495405297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005116_100 |
0.1500413149 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 15 |
Hb_005288_180 |
0.150509103 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_022425_050 |
0.1513815046 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 17 |
Hb_002447_050 |
0.1531368634 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000254_110 |
0.1538297682 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic-like [Jatropha curcas] |
| 19 |
Hb_000665_070 |
0.1539101634 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_013968_010 |
0.155329668 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |