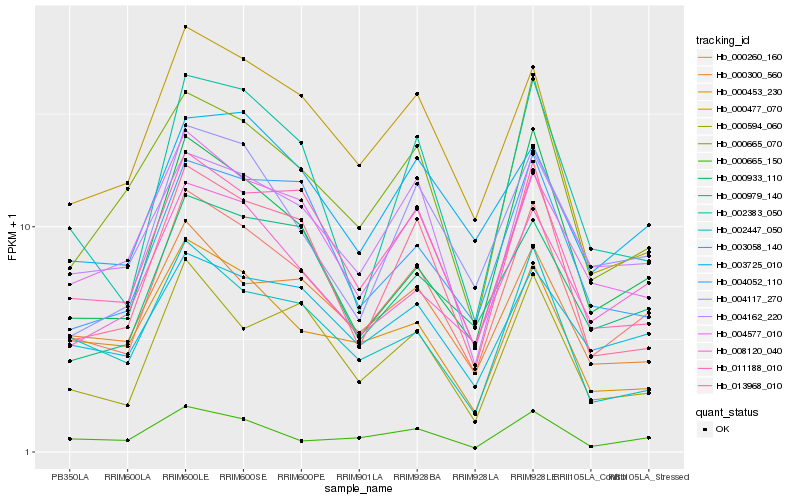
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_160 |
0.0 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_011188_010 |
0.0962367424 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008120_040 |
0.0963244562 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 4 |
Hb_000933_110 |
0.0969853173 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 5 |
Hb_004577_010 |
0.0978622197 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 6 |
Hb_013968_010 |
0.1022271029 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 7 |
Hb_004117_270 |
0.1026133651 |
- |
- |
- |
| 8 |
Hb_004162_220 |
0.1036165131 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000665_150 |
0.1039590956 |
- |
- |
heparanase, putative [Ricinus communis] |
| 10 |
Hb_000300_560 |
0.1054233939 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000594_060 |
0.1055756664 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 12 |
Hb_002383_050 |
0.1069327434 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 13 |
Hb_000665_070 |
0.1075206033 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002447_050 |
0.1091892521 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000979_140 |
0.1102766898 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 16 |
Hb_004052_110 |
0.1113302921 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003058_140 |
0.1115482032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000453_230 |
0.115323276 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_000477_070 |
0.1165904842 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 20 |
Hb_003725_010 |
0.1178158207 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |