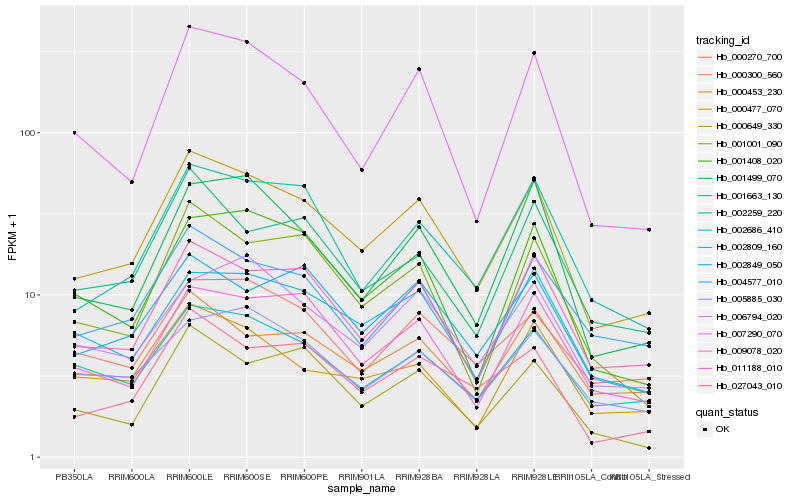
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000477_070 |
0.0 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 2 |
Hb_011188_010 |
0.0764617984 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002259_220 |
0.0931516505 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 4 |
Hb_027043_010 |
0.0943830675 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD4 [Jatropha curcas] |
| 5 |
Hb_002686_410 |
0.096960292 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_007290_070 |
0.0969850351 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 7 |
Hb_002809_160 |
0.1008626807 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 8 |
Hb_009078_020 |
0.1009760457 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 9 |
Hb_004577_010 |
0.1029331619 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 10 |
Hb_002849_050 |
0.1040166986 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 11 |
Hb_000300_560 |
0.1041984661 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001499_070 |
0.1064111348 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 13 |
Hb_000270_700 |
0.1084196347 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 14 |
Hb_001001_090 |
0.108422657 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 15 |
Hb_001408_020 |
0.1091001181 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 16 |
Hb_001663_130 |
0.1109789776 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 17 |
Hb_006794_020 |
0.1109999378 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 18 |
Hb_000649_330 |
0.111817181 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 19 |
Hb_000453_230 |
0.1132375835 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_005885_030 |
0.1140362198 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |