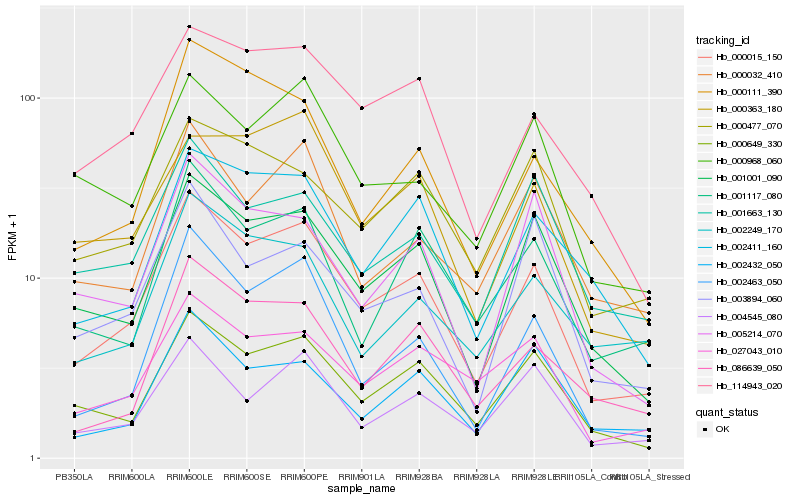
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000015_150 |
0.0 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 2 |
Hb_001001_090 |
0.0995583341 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 3 |
Hb_000649_330 |
0.100922563 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 4 |
Hb_114943_020 |
0.1120981222 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase 2 [Jatropha curcas] |
| 5 |
Hb_002463_050 |
0.1162753169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002411_160 |
0.1220831858 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 7 |
Hb_027043_010 |
0.1227369698 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD4 [Jatropha curcas] |
| 8 |
Hb_086639_050 |
0.1260742783 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004545_080 |
0.1288575298 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_001117_080 |
0.1288817596 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000968_060 |
0.1299763429 |
- |
- |
PREDICTED: gamma-interferon-inducible-lysosomal thiol reductase [Jatropha curcas] |
| 12 |
Hb_000363_180 |
0.1300660833 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 13 |
Hb_000032_410 |
0.1325739672 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 14 |
Hb_000477_070 |
0.1336585902 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 15 |
Hb_005214_070 |
0.1361861166 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 16 |
Hb_000111_390 |
0.1366678367 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001663_130 |
0.1374576728 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 18 |
Hb_002432_050 |
0.1385482022 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 19 |
Hb_002249_170 |
0.1386767889 |
- |
- |
copine, putative [Ricinus communis] |
| 20 |
Hb_003894_060 |
0.1398019242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |