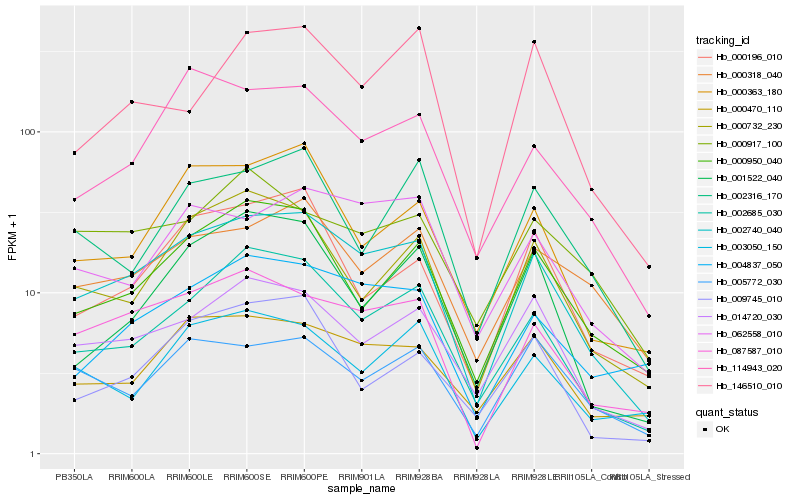
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002740_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s00350g [Populus trichocarpa] |
| 2 |
Hb_002685_030 |
0.1024084877 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 3 |
Hb_087587_010 |
0.1029807982 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_014720_030 |
0.1152623081 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 5 |
Hb_000470_110 |
0.1216955185 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |
| 6 |
Hb_000950_040 |
0.1248666092 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 7 |
Hb_114943_020 |
0.1253963791 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase 2 [Jatropha curcas] |
| 8 |
Hb_000732_230 |
0.1271377536 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000363_180 |
0.1272457237 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 10 |
Hb_062558_010 |
0.128060557 |
- |
- |
PREDICTED: L-2-hydroxyglutarate dehydrogenase, mitochondrial isoform X1 [Populus euphratica] |
| 11 |
Hb_004837_050 |
0.1292516577 |
- |
- |
- |
| 12 |
Hb_000318_040 |
0.1306756833 |
- |
- |
hypothetical protein JCGZ_04877 [Jatropha curcas] |
| 13 |
Hb_000196_010 |
0.138786992 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 14 |
Hb_003050_150 |
0.1416947491 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 15 |
Hb_005772_030 |
0.1422878942 |
- |
- |
PREDICTED: protein trichome birefringence-like 35 [Jatropha curcas] |
| 16 |
Hb_001522_040 |
0.144547541 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_002316_170 |
0.1483938926 |
- |
- |
purine permease, putative [Ricinus communis] |
| 18 |
Hb_000917_100 |
0.1511487896 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 19 |
Hb_009745_010 |
0.1517415388 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 20 |
Hb_146510_010 |
0.1533171188 |
- |
- |
cinnamyl alcohol dehydrogenase [Hevea brasiliensis] |