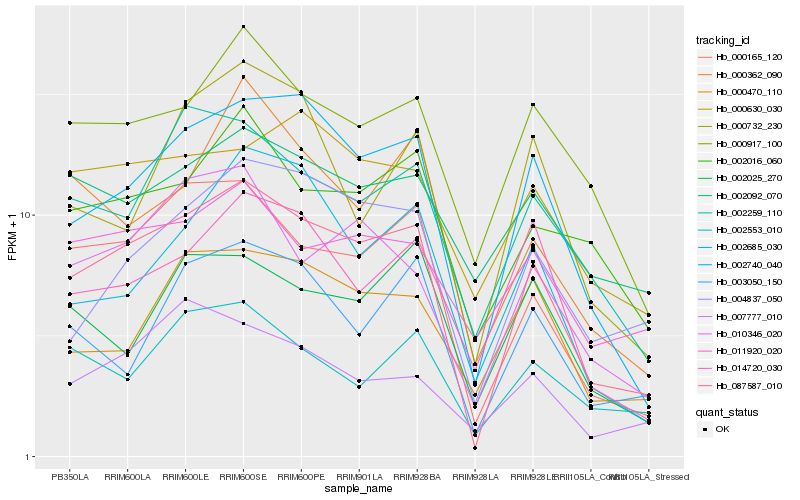
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_087587_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 2 |
Hb_002740_040 |
0.1029807982 |
- |
- |
hypothetical protein POPTR_0005s00350g [Populus trichocarpa] |
| 3 |
Hb_000165_120 |
0.107569567 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 4 |
Hb_000917_100 |
0.1373762889 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 5 |
Hb_010346_020 |
0.1381915701 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 6 |
Hb_002259_110 |
0.1388599809 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 7 |
Hb_004837_050 |
0.1424111597 |
- |
- |
- |
| 8 |
Hb_002016_060 |
0.1455538541 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 9 |
Hb_003050_150 |
0.1475666605 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |
| 10 |
Hb_014720_030 |
0.1526258249 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 11 |
Hb_000362_090 |
0.1533027542 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002553_010 |
0.1538640339 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 2-like [Populus euphratica] |
| 13 |
Hb_000470_110 |
0.1542825566 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |
| 14 |
Hb_011920_020 |
0.1544619355 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 15 |
Hb_002685_030 |
0.156713509 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 16 |
Hb_000630_030 |
0.1568778336 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 isoform X1 [Populus euphratica] |
| 17 |
Hb_007777_010 |
0.1580297621 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 18 |
Hb_002092_070 |
0.1596075958 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 19 |
Hb_002025_270 |
0.1609333422 |
- |
- |
PREDICTED: uncharacterized protein At4g15970 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000732_230 |
0.1609680807 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |