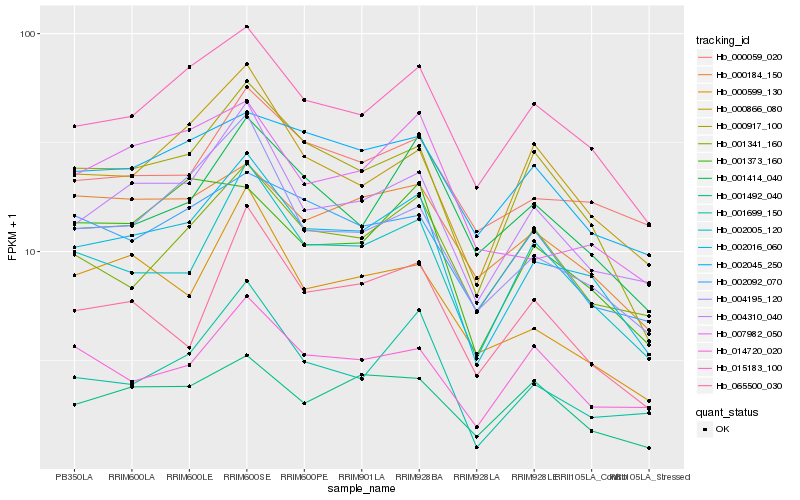
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 2 |
Hb_015183_100 |
0.081887784 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 3 |
Hb_004310_040 |
0.0838206842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000917_100 |
0.0842632598 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 5 |
Hb_002005_120 |
0.087611129 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 6 |
Hb_000866_080 |
0.1053710619 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000059_020 |
0.1057299272 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 8 |
Hb_004195_120 |
0.1065288023 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 9 |
Hb_001699_150 |
0.1077648523 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 10 |
Hb_001341_160 |
0.1112214114 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000184_150 |
0.1140474152 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 12 |
Hb_001414_040 |
0.1146597667 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002045_250 |
0.1164968601 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 14 |
Hb_002092_070 |
0.1181483525 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 15 |
Hb_007982_050 |
0.1183451423 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Jatropha curcas] |
| 16 |
Hb_001373_160 |
0.118416159 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 17 |
Hb_065500_030 |
0.1197804128 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 18 |
Hb_001492_040 |
0.1198714287 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_014720_020 |
0.120791868 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 20 |
Hb_000599_130 |
0.1208114426 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |