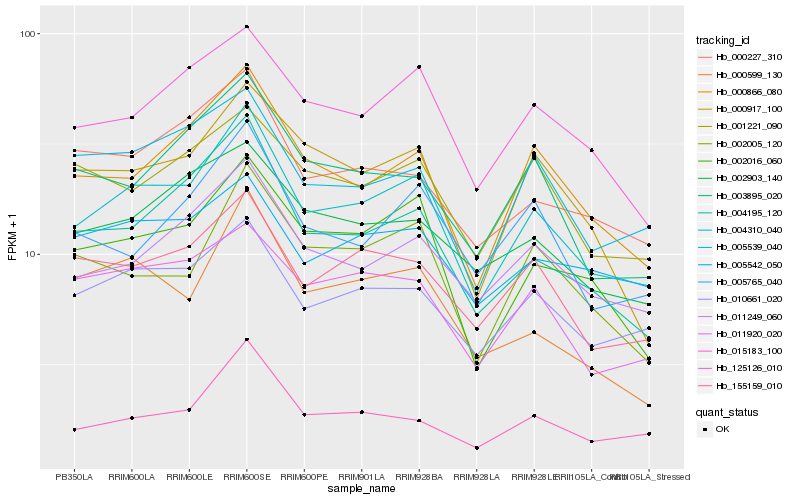
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004310_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002016_060 |
0.0838206842 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 3 |
Hb_000866_080 |
0.0849502078 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004195_120 |
0.0854224002 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 5 |
Hb_010661_020 |
0.0862759836 |
- |
- |
PREDICTED: uncharacterized protein LOC105630767 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011249_060 |
0.0878046508 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 7 |
Hb_015183_100 |
0.0898460482 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 8 |
Hb_125126_010 |
0.0938523781 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 9 |
Hb_155159_010 |
0.0940926293 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 10 |
Hb_011920_020 |
0.0979797562 |
- |
- |
PREDICTED: uncharacterized protein LOC105645397 [Jatropha curcas] |
| 11 |
Hb_002005_120 |
0.0994931648 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 12 |
Hb_005539_040 |
0.1009044556 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X3 [Jatropha curcas] |
| 13 |
Hb_002903_140 |
0.1028121887 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 14 |
Hb_000227_310 |
0.1038442206 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_003895_020 |
0.1044224646 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 16 |
Hb_005765_040 |
0.1045261489 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 17 |
Hb_000599_130 |
0.1062486207 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 18 |
Hb_000917_100 |
0.1065622704 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 19 |
Hb_001221_090 |
0.1074066014 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005542_050 |
0.1074259797 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable inactive serine/threonine-protein kinase scy1 [Prunus mume] |