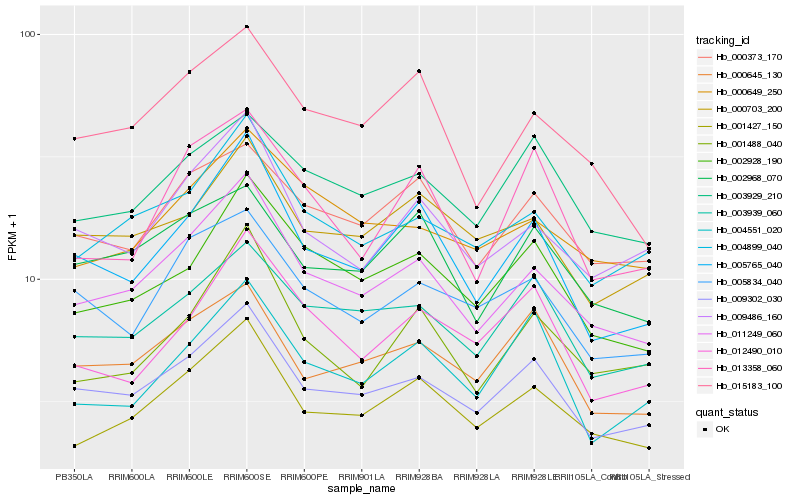
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001427_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637790 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011249_060 |
0.0562452825 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 3 |
Hb_004899_040 |
0.0813435213 |
transcription factor |
TF Family: ARF |
Auxin response factor [Medicago truncatula] |
| 4 |
Hb_009302_030 |
0.0832002662 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 5 |
Hb_001488_040 |
0.0833142471 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 6 |
Hb_002928_190 |
0.0852997816 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 7 |
Hb_005765_040 |
0.0882583286 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 8 |
Hb_000373_170 |
0.0887404952 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 9 |
Hb_009486_160 |
0.0911670832 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 10 |
Hb_003929_210 |
0.0945319875 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 11 |
Hb_015183_100 |
0.0946196847 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 12 |
Hb_000703_200 |
0.0952849146 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000645_130 |
0.09588399 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_003939_060 |
0.0963540098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000649_250 |
0.097748803 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_002968_070 |
0.0984758465 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 17 |
Hb_004551_020 |
0.1005357834 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 18 |
Hb_013358_060 |
0.1009037897 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 19 |
Hb_012490_010 |
0.1020262066 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005834_040 |
0.1030269287 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |