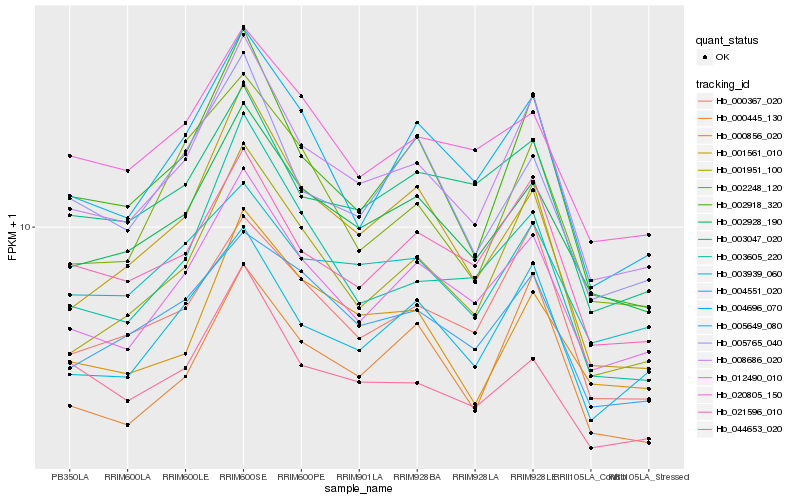
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008686_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002918_320 |
0.0613741866 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012490_010 |
0.0614227137 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001951_100 |
0.0659756989 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 5 |
Hb_002928_190 |
0.0662313875 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 6 |
Hb_004551_020 |
0.0684501666 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 7 |
Hb_000367_020 |
0.0751307006 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 8 |
Hb_004696_070 |
0.0759051134 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 9 |
Hb_021596_010 |
0.0774489729 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 10 |
Hb_003605_220 |
0.0856993948 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 11 |
Hb_005649_080 |
0.086877567 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 12 |
Hb_002248_120 |
0.0883667921 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 13 |
Hb_044653_020 |
0.0903523948 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 14 |
Hb_003047_020 |
0.0907966192 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_003939_060 |
0.0911262764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001561_010 |
0.0923335979 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000445_130 |
0.0925041031 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 18 |
Hb_000856_020 |
0.0927786951 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 19 |
Hb_005765_040 |
0.0929075006 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 20 |
Hb_020805_150 |
0.0931481349 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |