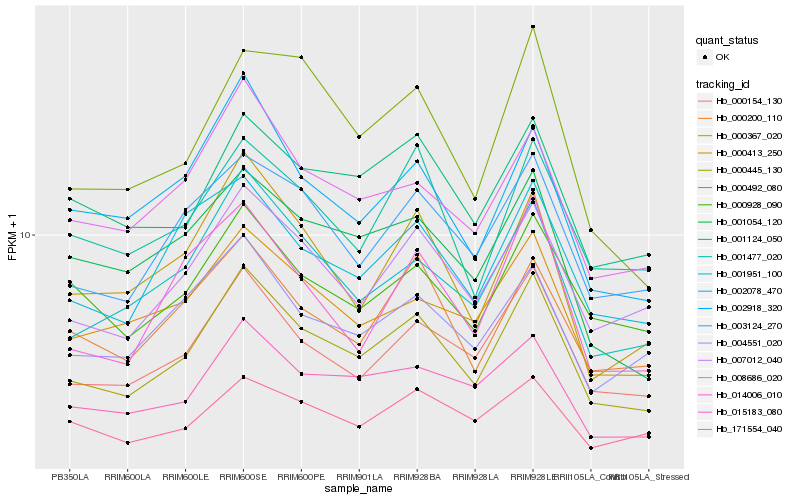
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000445_130 |
0.0 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_12159 [Jatropha curcas] |
| 2 |
Hb_000154_130 |
0.0716250851 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 3 |
Hb_007012_040 |
0.0784117241 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 4 |
Hb_001951_100 |
0.0824001319 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 5 |
Hb_015183_080 |
0.0868955415 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 6 |
Hb_003124_270 |
0.0877843718 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 7 |
Hb_000367_020 |
0.0920030855 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 8 |
Hb_008686_020 |
0.0925041031 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000413_250 |
0.0933912904 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 10 |
Hb_002078_470 |
0.0954152381 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000200_110 |
0.0971021572 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002918_320 |
0.0980002367 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000492_080 |
0.0995064035 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 [Jatropha curcas] |
| 14 |
Hb_001124_050 |
0.0997459339 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 15 |
Hb_001054_120 |
0.099817449 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 16 |
Hb_000928_090 |
0.1000974258 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 17 |
Hb_004551_020 |
0.1001715417 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 18 |
Hb_001477_020 |
0.100478676 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 19 |
Hb_014006_010 |
0.1013532093 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 20 |
Hb_171554_040 |
0.1035855409 |
- |
- |
PREDICTED: phytanoyl-CoA dioxygenase [Populus euphratica] |