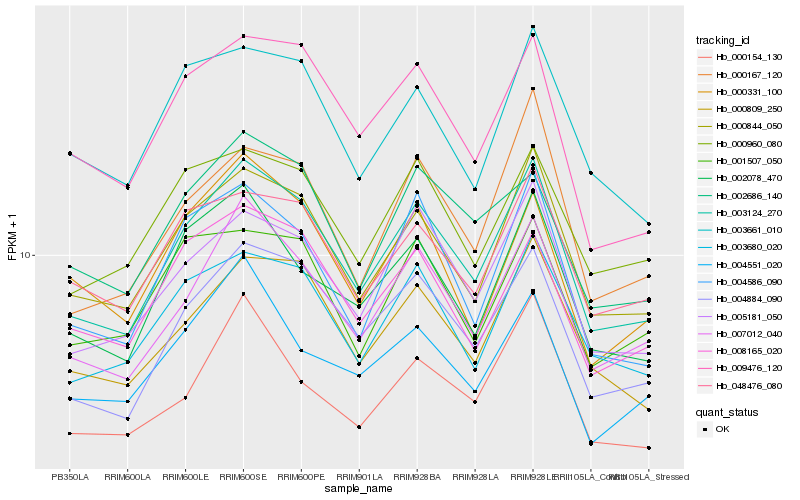
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_270 |
0.0 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 2 |
Hb_000844_050 |
0.0466768786 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 3 |
Hb_004884_090 |
0.0477622552 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 4 |
Hb_007012_040 |
0.0512690617 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 5 |
Hb_005181_050 |
0.0590452128 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 6 |
Hb_000809_250 |
0.0625370141 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 7 |
Hb_003680_020 |
0.0658349625 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002078_470 |
0.0699721004 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_009476_120 |
0.0724178691 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 10 |
Hb_004586_090 |
0.0725977857 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_048476_080 |
0.0739420887 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 12 |
Hb_008165_020 |
0.0748071771 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000960_080 |
0.0763743673 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000167_120 |
0.0766643769 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 15 |
Hb_002686_140 |
0.0783859014 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003661_010 |
0.0801313103 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001507_050 |
0.0822771853 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000331_100 |
0.0852290332 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004551_020 |
0.0862808247 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 20 |
Hb_000154_130 |
0.0868181469 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |