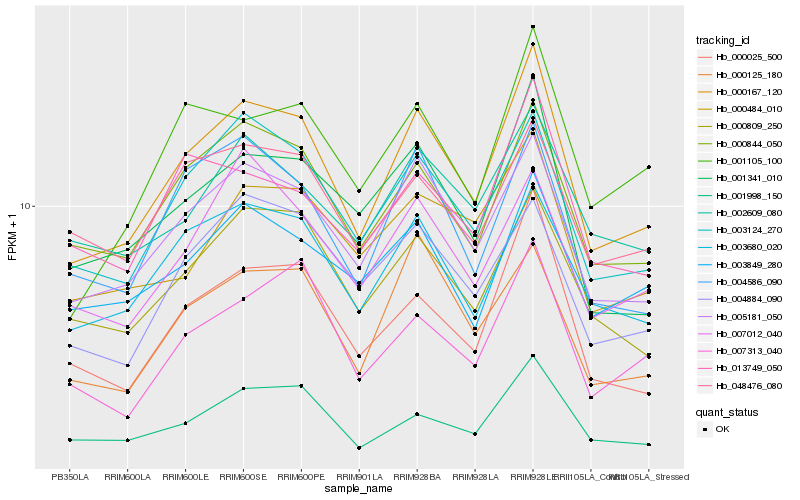
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000167_120 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 2 |
Hb_005181_050 |
0.0627098723 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 3 |
Hb_000844_050 |
0.075766183 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 4 |
Hb_003124_270 |
0.0766643769 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 5 |
Hb_000025_500 |
0.0829618197 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_001998_150 |
0.0830298424 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 7 |
Hb_003680_020 |
0.0850403819 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000809_250 |
0.0873941144 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 9 |
Hb_001341_010 |
0.087878315 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004586_090 |
0.0911970896 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000125_180 |
0.0954605239 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 12 |
Hb_001105_100 |
0.098454933 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 13 |
Hb_007313_040 |
0.0985197037 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 14 |
Hb_004884_090 |
0.098869538 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 15 |
Hb_003849_280 |
0.0991984431 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 16 |
Hb_007012_040 |
0.0994333704 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 17 |
Hb_000484_010 |
0.1006283546 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 23 [Jatropha curcas] |
| 18 |
Hb_002609_080 |
0.1022716566 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_013749_050 |
0.1033437887 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 20 |
Hb_048476_080 |
0.1033457294 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |