| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
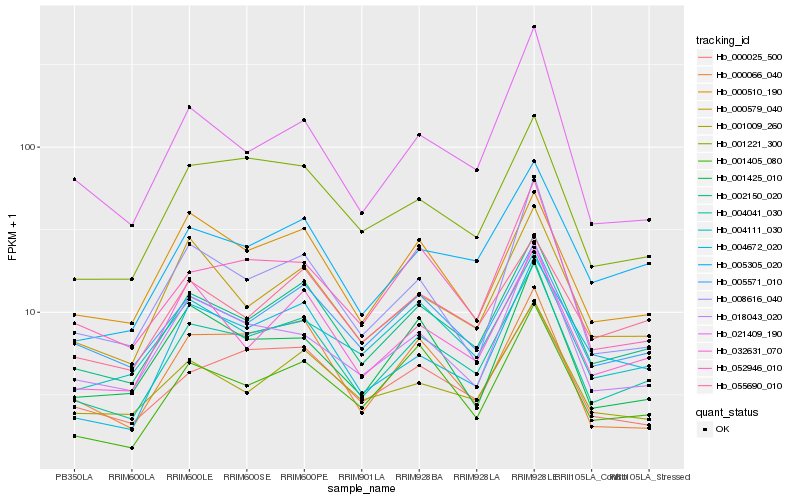
Hb_004041_030 |
0.0 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 2 |
Hb_002150_020 |
0.0480784771 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_005305_020 |
0.0818859303 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001221_300 |
0.088762101 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_052946_010 |
0.0911532364 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 6 |
Hb_000025_500 |
0.0923024284 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 7 |
Hb_001405_080 |
0.0956158124 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 8 |
Hb_032631_070 |
0.0964949565 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_005571_010 |
0.0965114011 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 10 |
Hb_000579_040 |
0.100826056 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 11 |
Hb_004111_030 |
0.1008591608 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000066_040 |
0.1015389042 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 13 |
Hb_001009_260 |
0.1016318873 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000510_190 |
0.1022481509 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 15 |
Hb_018043_020 |
0.1032190448 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_055690_010 |
0.1043365518 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_008616_040 |
0.1051405702 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 18 |
Hb_021409_190 |
0.1082987735 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 19 |
Hb_001425_010 |
0.1096678032 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 20 |
Hb_004672_020 |
0.1100934783 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |