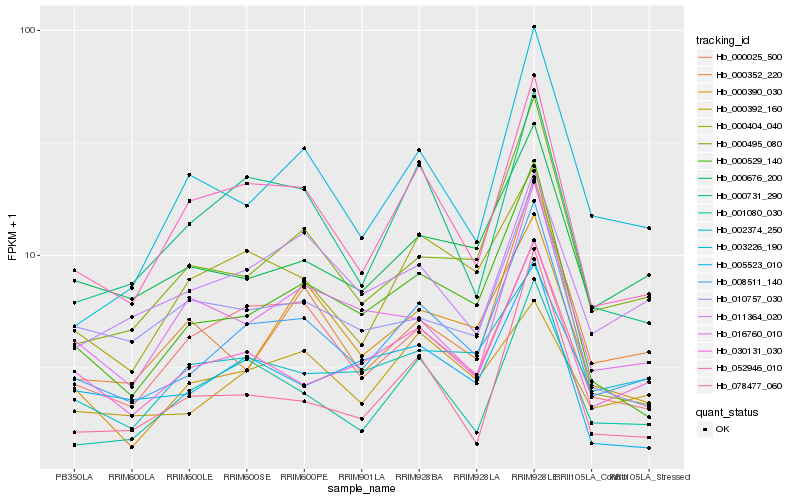
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008511_140 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 2 |
Hb_052946_010 |
0.0967731104 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 3 |
Hb_000529_140 |
0.1023554285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000495_080 |
0.1060923346 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 5 |
Hb_030131_030 |
0.1107824409 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g42310, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001080_030 |
0.1184454729 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 7 |
Hb_000676_200 |
0.1184897018 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 8 |
Hb_000731_290 |
0.1247258888 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_003226_190 |
0.1280976993 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 10 |
Hb_000025_500 |
0.1324174138 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 11 |
Hb_000352_220 |
0.1332172611 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000390_030 |
0.1360710234 |
- |
- |
hypothetical protein ZEAMMB73_364791 [Zea mays] |
| 13 |
Hb_078477_060 |
0.1366965174 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 14 |
Hb_005523_010 |
0.1393425767 |
transcription factor |
TF Family: PHD |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 15 |
Hb_016760_010 |
0.139494961 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_002374_250 |
0.1397985067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010757_030 |
0.1399270077 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_011364_020 |
0.140470957 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000392_160 |
0.1432040853 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000404_040 |
0.1438609004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |