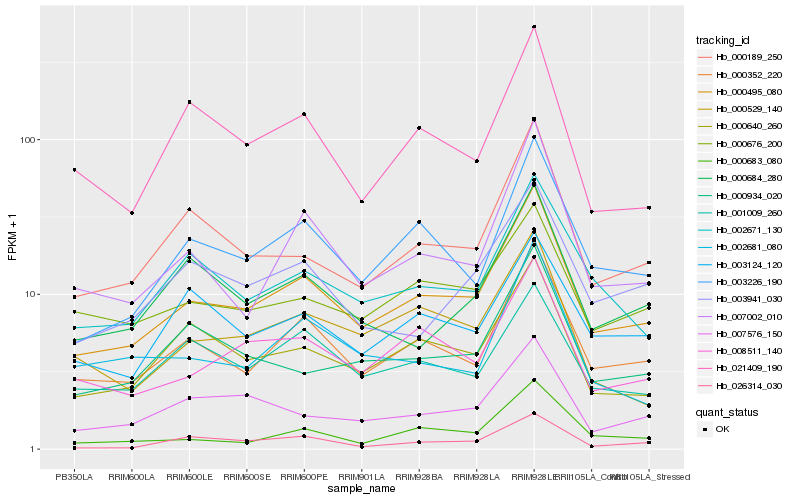
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000495_080 |
0.0 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 2 |
Hb_000352_220 |
0.0728063772 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003226_190 |
0.095457359 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 4 |
Hb_008511_140 |
0.1060923346 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 5 |
Hb_000189_250 |
0.106663795 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007002_010 |
0.1074457673 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 7 |
Hb_000640_260 |
0.1096760404 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002671_130 |
0.1151664507 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 9 |
Hb_000683_080 |
0.1167970267 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 10 |
Hb_000676_200 |
0.1220396071 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 11 |
Hb_000934_020 |
0.1236236785 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 12 |
Hb_007576_150 |
0.1263655687 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 13 |
Hb_021409_190 |
0.1264215197 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 14 |
Hb_003124_120 |
0.1273989752 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 15 |
Hb_000529_140 |
0.1276153829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003941_030 |
0.1276720057 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 17 |
Hb_000684_280 |
0.1284208651 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |
| 18 |
Hb_026314_030 |
0.1308130644 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 19 |
Hb_002681_080 |
0.1334639883 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001009_260 |
0.1335707239 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |