| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
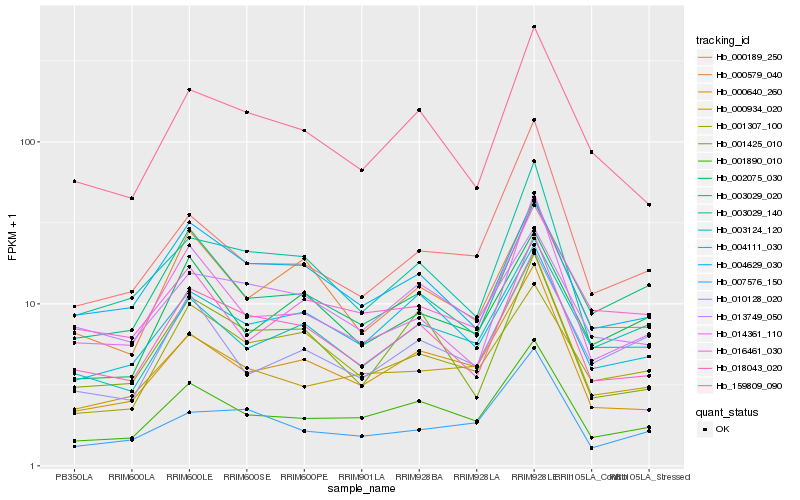
Hb_001890_010 |
0.0 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 2 |
Hb_000640_260 |
0.0780528268 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004111_030 |
0.0860681111 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_003124_120 |
0.0921521298 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 5 |
Hb_018043_020 |
0.0963695058 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000934_020 |
0.1056551857 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 7 |
Hb_159809_090 |
0.1091535331 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 8 |
Hb_010128_020 |
0.1121835318 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 9 |
Hb_016461_030 |
0.1184786443 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_003029_020 |
0.1185356386 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 11 |
Hb_002075_030 |
0.1189847248 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003029_140 |
0.1196403588 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004629_030 |
0.1206358963 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_007576_150 |
0.1214162159 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g57430, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000579_040 |
0.1215118666 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 16 |
Hb_013749_050 |
0.1227113302 |
- |
- |
PREDICTED: uncharacterized protein LOC105647765 isoform X2 [Jatropha curcas] |
| 17 |
Hb_014361_110 |
0.1250450984 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000189_250 |
0.1251292829 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001425_010 |
0.1254912799 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 20 |
Hb_001307_100 |
0.1262202423 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |