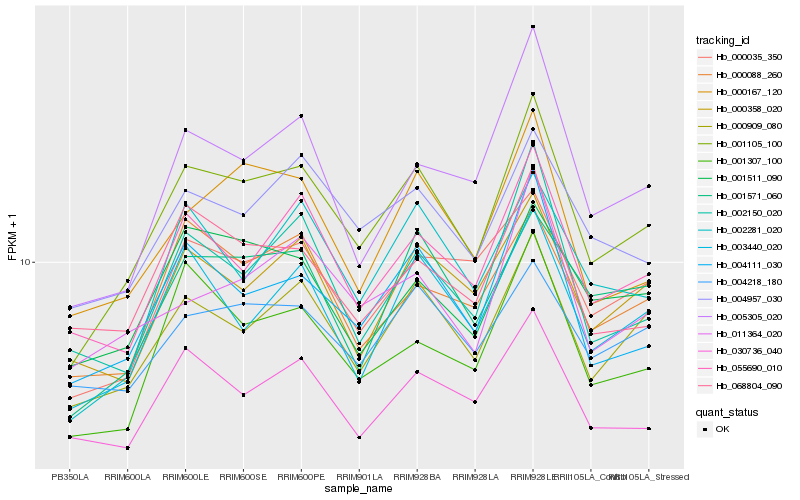
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001105_100 |
0.0 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 2 |
Hb_000909_080 |
0.0797662178 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 3 |
Hb_004111_030 |
0.0866087531 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_055690_010 |
0.0944072042 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003440_020 |
0.0965701517 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000167_120 |
0.098454933 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 7 |
Hb_001571_060 |
0.0987302035 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 8 |
Hb_004957_030 |
0.0995317912 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 9 |
Hb_000035_350 |
0.1008338689 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001307_100 |
0.1020505973 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_000358_020 |
0.102768303 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 12 |
Hb_002281_020 |
0.1038955822 |
- |
- |
PREDICTED: actin-related protein 8 [Jatropha curcas] |
| 13 |
Hb_002150_020 |
0.1045285824 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_030736_040 |
0.1049791902 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005305_020 |
0.1068564498 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000088_260 |
0.1083480608 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 17 |
Hb_001511_090 |
0.1085155422 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 18 |
Hb_068804_090 |
0.1087910579 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 19 |
Hb_011364_020 |
0.1088089827 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004218_180 |
0.1115653005 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |