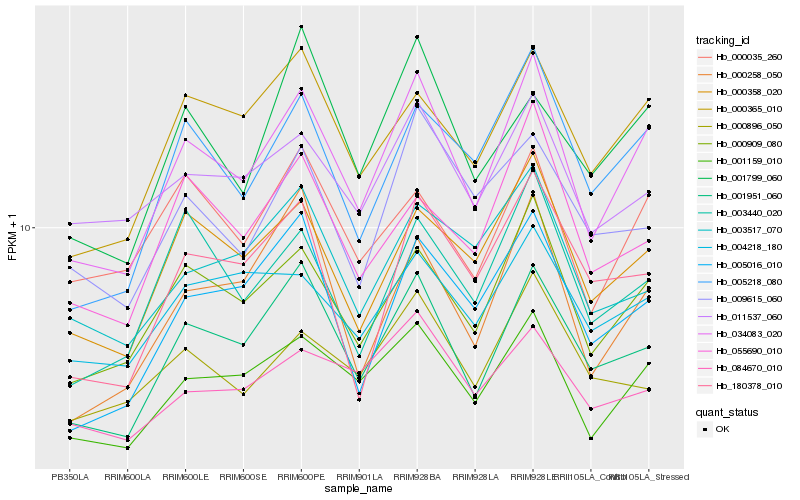
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034083_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000909_080 |
0.0664845644 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 3 |
Hb_001159_010 |
0.0714973989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001951_060 |
0.0819910457 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 5 |
Hb_000358_020 |
0.0926384149 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 6 |
Hb_005218_080 |
0.0927226504 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000365_010 |
0.0986017686 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 8 |
Hb_003440_020 |
0.1058924401 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_011537_060 |
0.1104347319 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 10 |
Hb_000035_260 |
0.1114618174 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 11 |
Hb_005016_010 |
0.1124181237 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 12 |
Hb_000258_050 |
0.1140545416 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_055690_010 |
0.1147330913 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004218_180 |
0.1174687273 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 15 |
Hb_000896_050 |
0.1178419343 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 16 |
Hb_180378_010 |
0.1181577004 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 17 |
Hb_084670_010 |
0.1197893248 |
- |
- |
PREDICTED: thiamine biosynthetic bifunctional enzyme TH1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_009615_060 |
0.1206165069 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 19 |
Hb_003517_070 |
0.1208789656 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 20 |
Hb_001799_060 |
0.12186491 |
- |
- |
Rab6 [Hevea brasiliensis] |