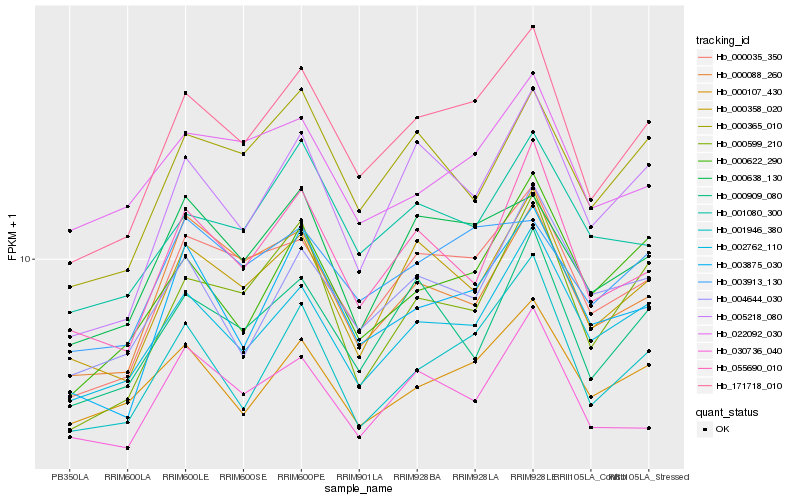
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171718_010 |
0.0 |
- |
- |
PREDICTED: disease resistance protein At4g27190-like [Eucalyptus grandis] |
| 2 |
Hb_000035_350 |
0.0622988932 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000358_020 |
0.0811151005 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 4 |
Hb_002762_110 |
0.0850107769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001946_380 |
0.085012314 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 6 |
Hb_003875_030 |
0.0855272203 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 7 |
Hb_005218_080 |
0.0872531956 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_055690_010 |
0.091766344 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000622_290 |
0.0975166261 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 10 |
Hb_000638_130 |
0.0979676534 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 11 |
Hb_000599_210 |
0.0980814598 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 12 |
Hb_001080_300 |
0.0982610368 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 13 |
Hb_000909_080 |
0.0996387374 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 14 |
Hb_000365_010 |
0.1017164652 |
- |
- |
hypothetical protein POPTR_0006s10920g [Populus trichocarpa] |
| 15 |
Hb_003913_130 |
0.1017336005 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 16 |
Hb_000107_430 |
0.1035172147 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 17 |
Hb_000088_260 |
0.1047423622 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 18 |
Hb_004644_030 |
0.1061735847 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 19 |
Hb_030736_040 |
0.1077259851 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 20 |
Hb_022092_030 |
0.1096081783 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |