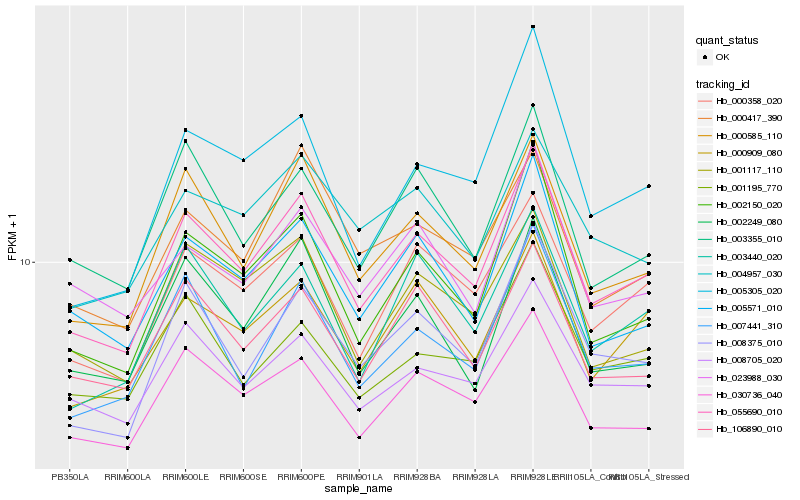
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055690_010 |
0.0 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000585_110 |
0.0594954651 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 3 |
Hb_030736_040 |
0.061360663 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000358_020 |
0.063096957 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 5 |
Hb_002150_020 |
0.0647612424 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_008375_010 |
0.0700914828 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 7 |
Hb_005571_010 |
0.0720659015 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 8 |
Hb_000909_080 |
0.0730912117 |
- |
- |
PREDICTED: inosine-5'-monophosphate dehydrogenase 2-like [Jatropha curcas] |
| 9 |
Hb_008705_020 |
0.0734404645 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_007441_310 |
0.0757610466 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_106890_010 |
0.0777037037 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_001117_110 |
0.0802895559 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 13 |
Hb_003355_010 |
0.0805720308 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 14 |
Hb_003440_020 |
0.0808234171 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_002249_080 |
0.0838060387 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 16 |
Hb_004957_030 |
0.0869820453 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 17 |
Hb_023988_030 |
0.0870045786 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 18 |
Hb_001195_770 |
0.0878122438 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 19 |
Hb_000417_390 |
0.0881124671 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 20 |
Hb_005305_020 |
0.0886033298 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |