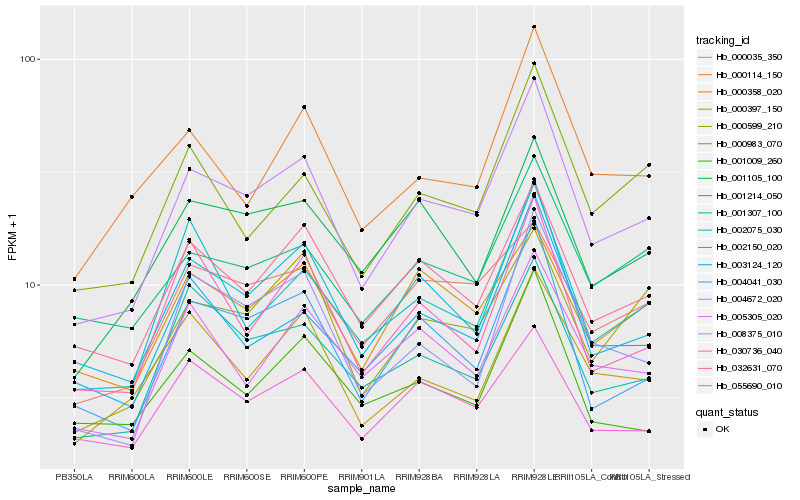
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005305_020 |
0.0 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002150_020 |
0.0753346983 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_004041_030 |
0.0818859303 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 4 |
Hb_055690_010 |
0.0886033298 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003124_120 |
0.0952533119 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 6 |
Hb_004672_020 |
0.1009136911 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 7 |
Hb_001307_100 |
0.1014359297 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 8 |
Hb_001214_050 |
0.1032881342 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000114_150 |
0.1033597797 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_000983_070 |
0.1035483052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008375_010 |
0.1056054101 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 12 |
Hb_000599_210 |
0.1060696238 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 13 |
Hb_032631_070 |
0.1066685992 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_001105_100 |
0.1068564498 |
- |
- |
PREDICTED: cathepsin B [Jatropha curcas] |
| 15 |
Hb_000397_150 |
0.109118636 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 16 |
Hb_000035_350 |
0.109486733 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_030736_040 |
0.1096942779 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000358_020 |
0.1097403641 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 19 |
Hb_002075_030 |
0.1107048383 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001009_260 |
0.1131047525 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |