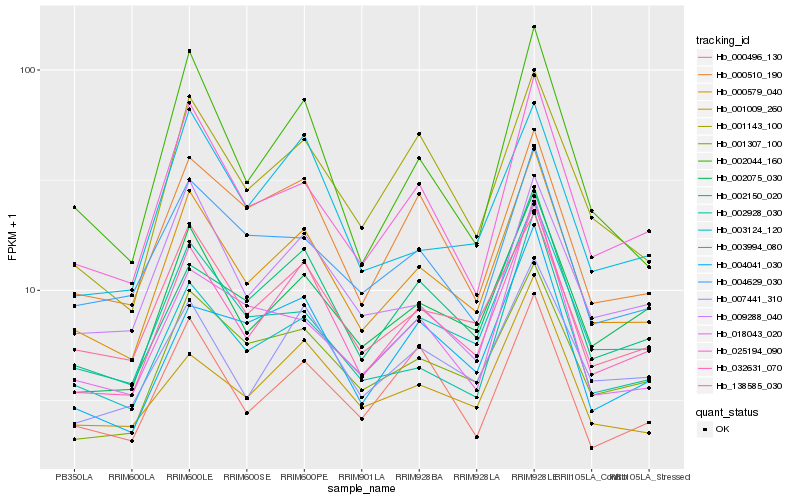
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_040 |
0.0 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 2 |
Hb_032631_070 |
0.0484034532 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_025194_090 |
0.075089514 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 4 |
Hb_002075_030 |
0.0766619948 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_138585_030 |
0.0789175694 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 6 |
Hb_001307_100 |
0.0836993364 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 7 |
Hb_001143_100 |
0.0863252439 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 8 |
Hb_002150_020 |
0.0865743369 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_002044_160 |
0.0891009779 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 10 |
Hb_007441_310 |
0.0907337036 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004629_030 |
0.0937481209 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_001009_260 |
0.0948379723 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002928_030 |
0.0963389918 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 14 |
Hb_003124_120 |
0.0972662015 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 15 |
Hb_000510_190 |
0.097286125 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 16 |
Hb_000496_130 |
0.0987122622 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 17 |
Hb_009288_040 |
0.0988365828 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 18 |
Hb_003994_080 |
0.100135125 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 19 |
Hb_018043_020 |
0.1001461515 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004041_030 |
0.100826056 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |