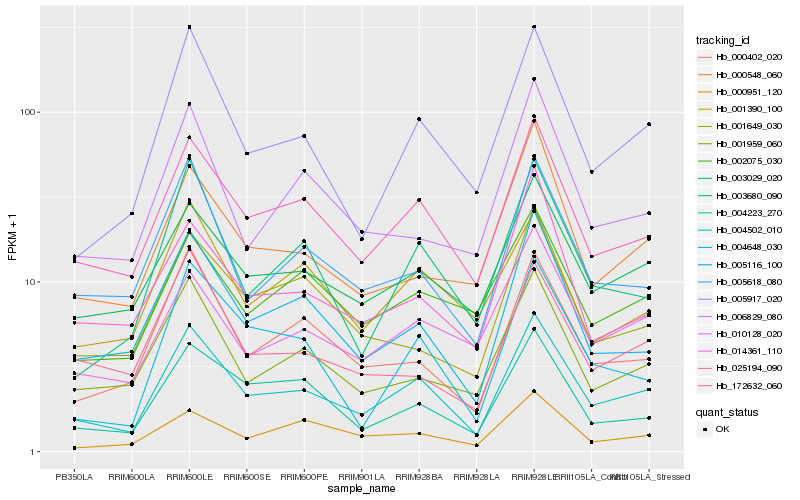
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004502_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 2 |
Hb_025194_090 |
0.0818405285 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 3 |
Hb_005917_020 |
0.1036431906 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 4 |
Hb_003029_020 |
0.1120307337 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 5 |
Hb_005116_100 |
0.1120891366 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 6 |
Hb_005618_080 |
0.1193102104 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000402_020 |
0.1208782439 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001959_060 |
0.1249714235 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 9 |
Hb_004223_270 |
0.1257665857 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003680_090 |
0.1274242364 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 11 |
Hb_010128_020 |
0.130503634 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 12 |
Hb_001390_100 |
0.1355557999 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000548_060 |
0.1360145813 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 14 |
Hb_004648_030 |
0.1362945667 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 15 |
Hb_002075_030 |
0.1371586615 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_172632_060 |
0.1399605729 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006829_080 |
0.1406560178 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 18 |
Hb_000951_120 |
0.1406723334 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 19 |
Hb_001649_030 |
0.141530524 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 20 |
Hb_014361_110 |
0.1416408425 |
- |
- |
RNA binding protein, putative [Ricinus communis] |