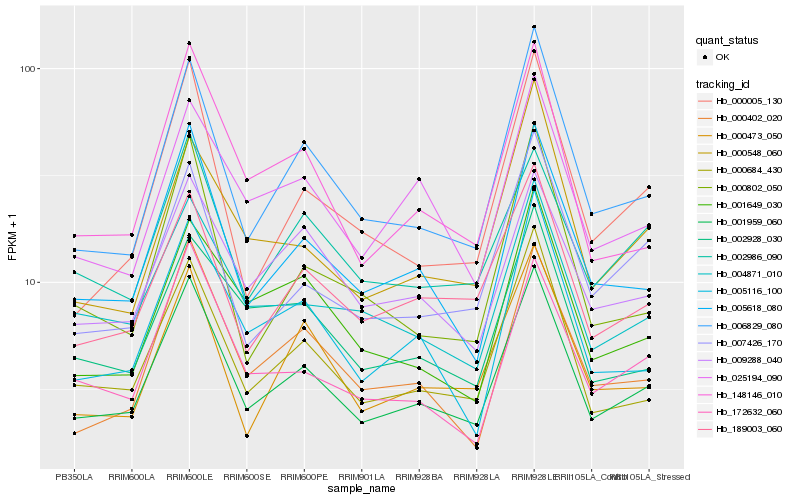
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_060 |
0.0 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006829_080 |
0.0700277041 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 3 |
Hb_005618_080 |
0.0700560785 |
- |
- |
PREDICTED: protoporphyrinogen oxidase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000684_430 |
0.0723177696 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g02830, chloroplastic [Jatropha curcas] |
| 5 |
Hb_148146_010 |
0.0881846207 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 6 |
Hb_189003_060 |
0.0895357521 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000402_020 |
0.0935072173 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_172632_060 |
0.0961687674 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005116_100 |
0.0964414414 |
- |
- |
PREDICTED: uncharacterized protein LOC105649999 [Jatropha curcas] |
| 10 |
Hb_004871_010 |
0.0965855821 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 11 |
Hb_007426_170 |
0.0975445679 |
transcription factor |
TF Family: TCP |
- |
| 12 |
Hb_000473_050 |
0.0976647263 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 13 |
Hb_000005_130 |
0.0993715934 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 14 |
Hb_009288_040 |
0.1028316638 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 15 |
Hb_025194_090 |
0.1048926065 |
- |
- |
coproporphyrinogen III oxidase, putative [Ricinus communis] |
| 16 |
Hb_000802_050 |
0.1062758848 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |
| 17 |
Hb_002986_090 |
0.1063585616 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_001649_030 |
0.1065386334 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002928_030 |
0.1068704289 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 20 |
Hb_000548_060 |
0.107781541 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |