| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
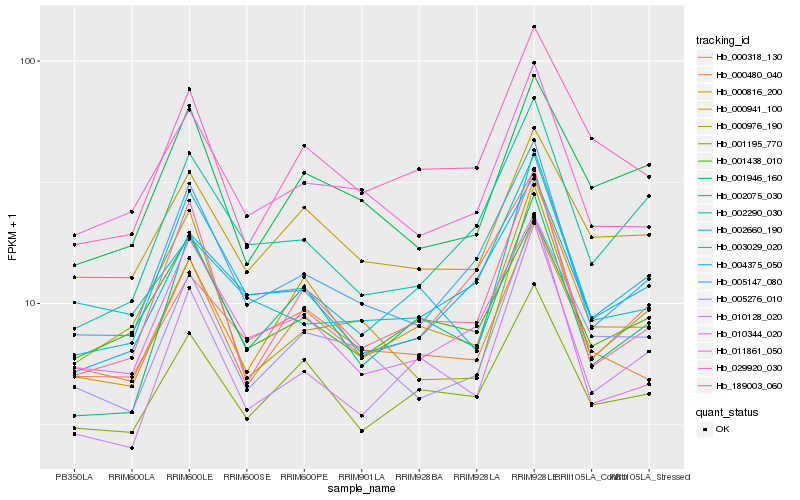
Hb_005147_080 |
0.0 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_002290_030 |
0.0786559305 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004375_050 |
0.0807545269 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_010344_020 |
0.084490401 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 5 |
Hb_011861_050 |
0.0880282286 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_189003_060 |
0.0881547474 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000318_130 |
0.1023308518 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_005276_010 |
0.1040540476 |
- |
- |
hypothetical protein CICLE_v10021605mg [Citrus clementina] |
| 9 |
Hb_002660_190 |
0.1062187954 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 10 |
Hb_001195_770 |
0.1063974791 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 11 |
Hb_000480_040 |
0.1065193764 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 12 |
Hb_002075_030 |
0.1068049392 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001438_010 |
0.108774663 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003029_020 |
0.1119360331 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 15 |
Hb_010128_020 |
0.1167320444 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 16 |
Hb_000816_200 |
0.1185636272 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 17 |
Hb_000941_100 |
0.1193125368 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 18 |
Hb_001946_160 |
0.1197625371 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 19 |
Hb_000976_190 |
0.1198354931 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 20 |
Hb_029920_030 |
0.1207006779 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |