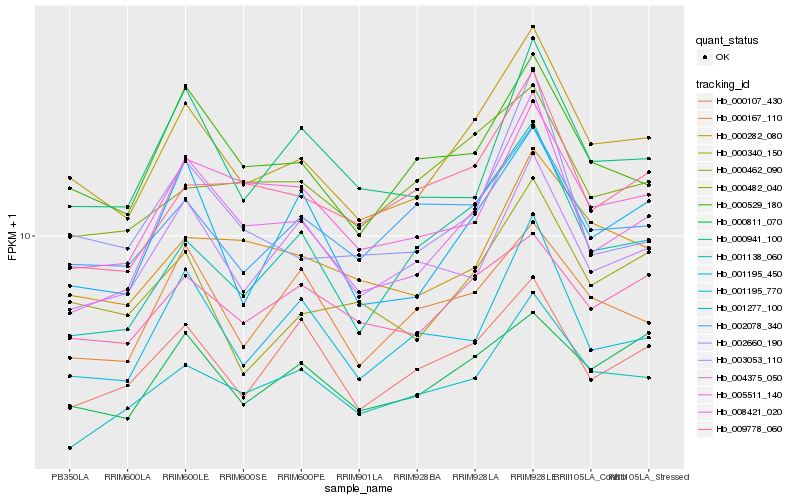
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 2 |
Hb_004375_050 |
0.088584212 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000529_180 |
0.0899860736 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000462_090 |
0.0946343733 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001277_100 |
0.0967825212 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 6 |
Hb_000811_070 |
0.0980662806 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 7 |
Hb_002078_340 |
0.0985486064 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001195_450 |
0.0996677921 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 9 |
Hb_000282_080 |
0.0998257188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001138_060 |
0.1017009394 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 11 |
Hb_008421_020 |
0.1019164708 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005511_140 |
0.1023979234 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 13 |
Hb_009778_060 |
0.1034426933 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 14 |
Hb_001195_770 |
0.1036751567 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 15 |
Hb_002660_190 |
0.1045422771 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 16 |
Hb_000941_100 |
0.1045937277 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 17 |
Hb_000107_430 |
0.1075045071 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 18 |
Hb_000167_110 |
0.1078905545 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000482_040 |
0.1096721932 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 20 |
Hb_003053_110 |
0.1098096386 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |