| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
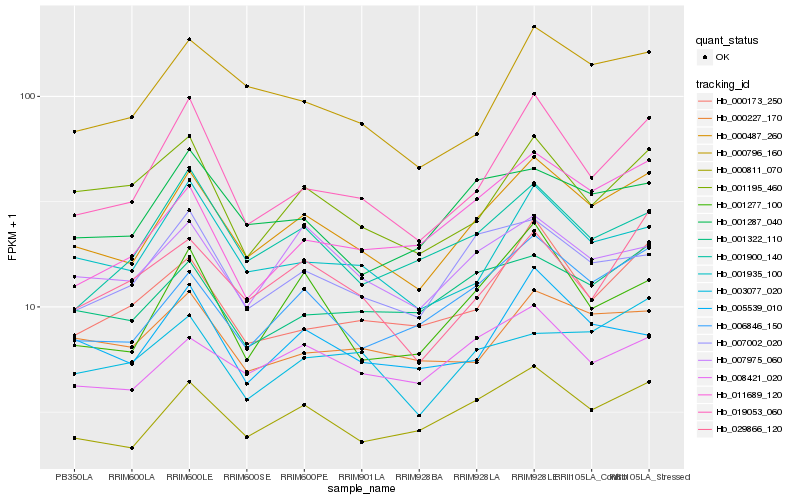
Hb_000487_260 |
0.0 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000811_070 |
0.0634211247 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 3 |
Hb_006846_150 |
0.0737641442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_019053_060 |
0.0797596552 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 5 |
Hb_000796_160 |
0.0876368884 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001935_100 |
0.0896685498 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 7 |
Hb_008421_020 |
0.0909357912 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007975_060 |
0.0930074455 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 9 |
Hb_007002_020 |
0.0933822849 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 10 |
Hb_001322_110 |
0.0957323468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001277_100 |
0.0957645269 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 12 |
Hb_001287_040 |
0.0961911352 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 13 |
Hb_000173_250 |
0.098834226 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 14 |
Hb_000227_170 |
0.0989649248 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 15 |
Hb_001195_460 |
0.0990929009 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_011689_120 |
0.1000924531 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 17 |
Hb_029866_120 |
0.1002416278 |
- |
- |
hypothetical protein CICLE_v10006070mg [Citrus clementina] |
| 18 |
Hb_005539_010 |
0.1012667856 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 19 |
Hb_003077_020 |
0.101784044 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_001900_140 |
0.1020677531 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |