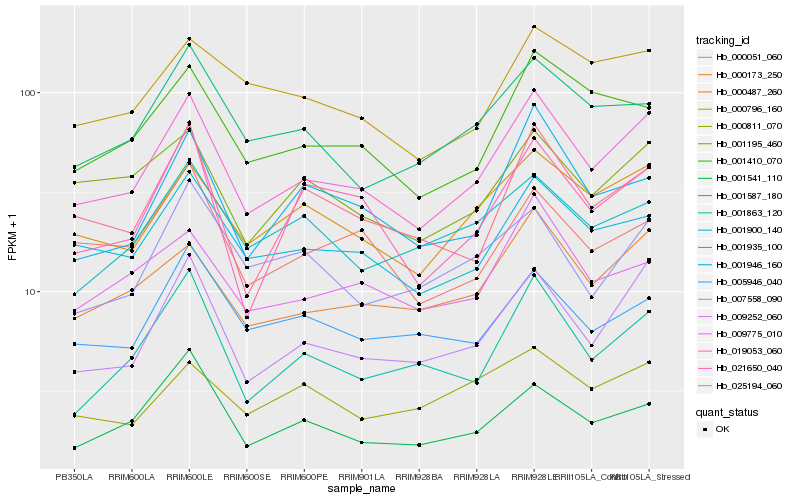
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019053_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 2 |
Hb_009252_060 |
0.073407203 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 3 |
Hb_000487_260 |
0.0797596552 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000173_250 |
0.080129302 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 5 |
Hb_001935_100 |
0.0844904975 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 6 |
Hb_001410_070 |
0.0965331388 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 7 |
Hb_001946_160 |
0.0971440931 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 8 |
Hb_025194_060 |
0.0973706548 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_001863_120 |
0.0987793686 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001195_460 |
0.0995457353 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_001587_180 |
0.1002744244 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 12 |
Hb_009775_010 |
0.1019011245 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 13 |
Hb_021650_040 |
0.1032712068 |
- |
- |
EG2771 [Manihot esculenta] |
| 14 |
Hb_001541_110 |
0.1060931884 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_005946_040 |
0.1061602034 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000796_160 |
0.1062287945 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000051_060 |
0.1079368132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001900_140 |
0.1084250692 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_007558_090 |
0.1099703478 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000811_070 |
0.1136636256 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |