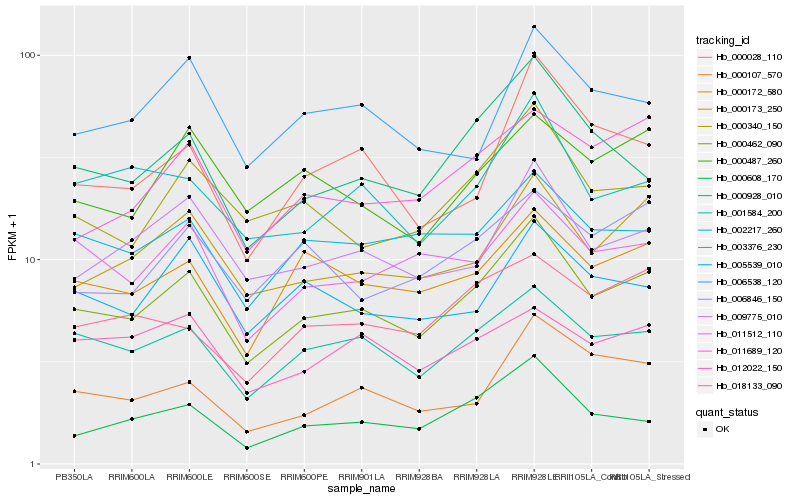
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000462_090 |
0.0 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001584_200 |
0.0761406965 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 3 |
Hb_000173_250 |
0.0891461151 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 4 |
Hb_000340_150 |
0.0946343733 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 5 |
Hb_000172_580 |
0.0946924534 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_009775_010 |
0.0949606487 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 7 |
Hb_003376_230 |
0.0979409826 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 8 |
Hb_002217_260 |
0.1002683228 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 9 |
Hb_000928_010 |
0.1011242299 |
- |
- |
Cell division protein ftsZ, putative [Ricinus communis] |
| 10 |
Hb_011512_110 |
0.1020577785 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000608_170 |
0.10499606 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 12 |
Hb_000487_260 |
0.1075919459 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 13 |
Hb_011689_120 |
0.1097607702 |
- |
- |
PREDICTED: uncharacterized protein At4g15545 [Jatropha curcas] |
| 14 |
Hb_005539_010 |
0.1103534995 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 15 |
Hb_018133_090 |
0.1126758689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006538_120 |
0.1128750056 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000028_110 |
0.112901896 |
- |
- |
PREDICTED: uncharacterized protein LOC105645305 [Jatropha curcas] |
| 18 |
Hb_006846_150 |
0.1129523074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000107_570 |
0.1146372258 |
- |
- |
PREDICTED: uncharacterized protein LOC105634044 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012022_150 |
0.115648881 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |