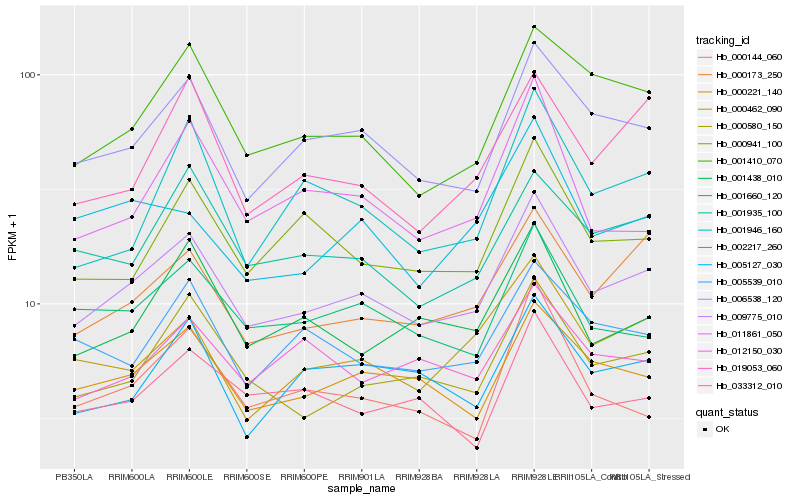
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009775_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 2 |
Hb_006538_120 |
0.0689110469 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000221_140 |
0.077574298 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000580_150 |
0.0801561306 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000173_250 |
0.0820030153 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 6 |
Hb_001410_070 |
0.0890844648 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 7 |
Hb_001660_120 |
0.0928966165 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001935_100 |
0.0930529976 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 9 |
Hb_001438_010 |
0.093952196 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000941_100 |
0.0942748171 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 11 |
Hb_005127_030 |
0.094669156 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 12 |
Hb_000462_090 |
0.0949606487 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005539_010 |
0.0981925774 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 14 |
Hb_012150_030 |
0.101057024 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_033312_010 |
0.1013926999 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_019053_060 |
0.1019011245 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 17 |
Hb_011861_050 |
0.1054753124 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_001946_160 |
0.1065663277 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 19 |
Hb_002217_260 |
0.106682811 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 20 |
Hb_000144_060 |
0.1073774003 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |