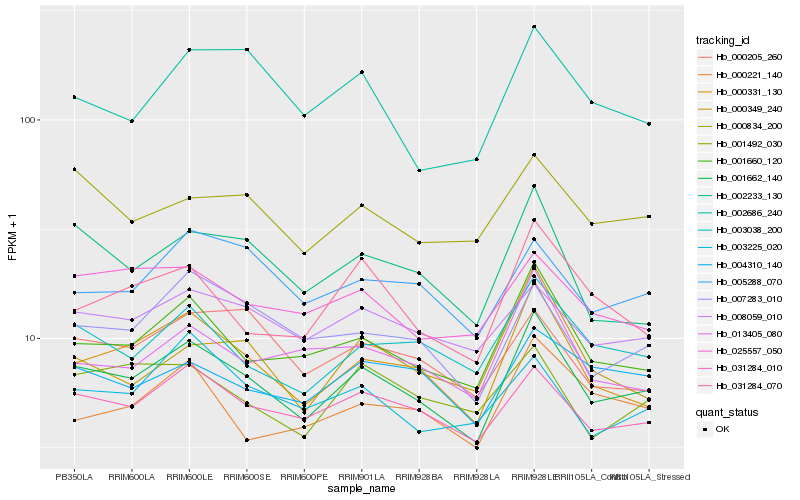
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001662_140 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000331_130 |
0.0693086024 |
transcription factor |
TF Family: Trihelix |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001660_120 |
0.0706627647 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_031284_010 |
0.0788477972 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 5 |
Hb_003038_200 |
0.0834974123 |
desease resistance |
Gene Name: MMR_HSR1 |
PREDICTED: GTPase Era, mitochondrial [Jatropha curcas] |
| 6 |
Hb_002233_130 |
0.0896234594 |
- |
- |
transporter, putative [Ricinus communis] |
| 7 |
Hb_008059_010 |
0.0912485413 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 8 |
Hb_007283_010 |
0.0935652547 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 9 |
Hb_000834_200 |
0.0946209213 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 10 |
Hb_031284_070 |
0.0961892753 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_025557_050 |
0.0978497117 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74630 [Jatropha curcas] |
| 12 |
Hb_004310_140 |
0.0987994875 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_005288_070 |
0.0994202506 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_013405_080 |
0.0997434853 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 15 |
Hb_001492_030 |
0.1015772092 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18975, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000205_260 |
0.1031288971 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 17 |
Hb_000221_140 |
0.1033270419 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_003225_020 |
0.1035114969 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 19 |
Hb_002686_240 |
0.1038797208 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 20 |
Hb_000349_240 |
0.1039617939 |
- |
- |
PREDICTED: probable aminotransferase ACS10 isoform X1 [Jatropha curcas] |