| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
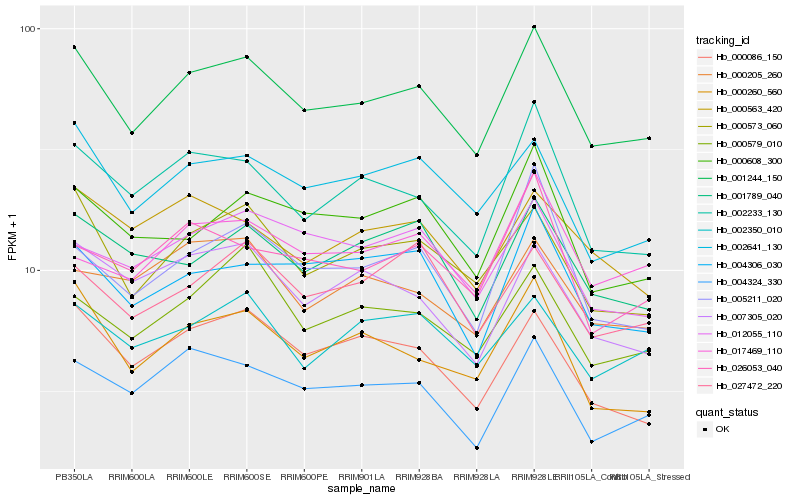
Hb_001244_150 |
0.0 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 2 |
Hb_000573_060 |
0.0649166193 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 3 |
Hb_002641_130 |
0.0696612268 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 4 |
Hb_002233_130 |
0.0711161375 |
- |
- |
transporter, putative [Ricinus communis] |
| 5 |
Hb_004324_330 |
0.0771134507 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 6 |
Hb_002350_010 |
0.0782359676 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 7 |
Hb_000608_300 |
0.0785068418 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 8 |
Hb_001789_040 |
0.079929594 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000086_150 |
0.0804599256 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004306_030 |
0.0808394423 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 11 |
Hb_000260_560 |
0.0812763 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000579_010 |
0.0822779477 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 13 |
Hb_027472_220 |
0.0857516647 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_007305_020 |
0.0861124296 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 15 |
Hb_012055_110 |
0.0864417228 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 16 |
Hb_017469_110 |
0.0869868402 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 17 |
Hb_005211_020 |
0.0870519953 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000563_420 |
0.0878484674 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 19 |
Hb_026053_040 |
0.0881169288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000205_260 |
0.0888760363 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |