| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
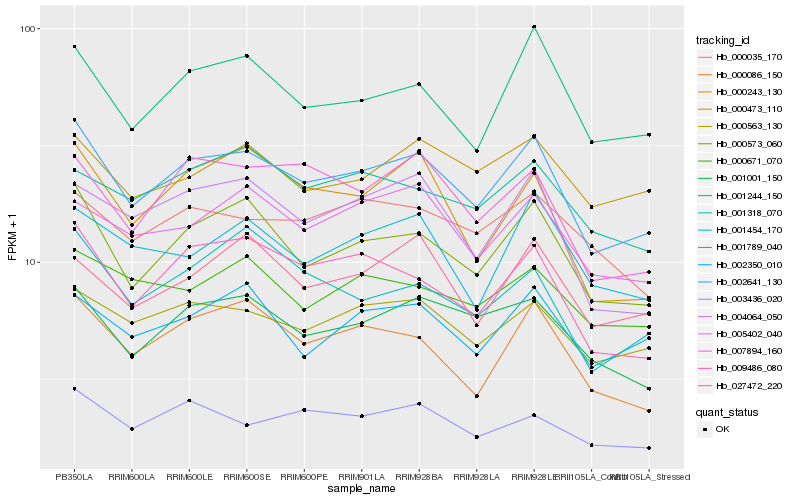
Hb_002641_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 2 |
Hb_000573_060 |
0.0520884627 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 3 |
Hb_001244_150 |
0.0696612268 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 4 |
Hb_000563_130 |
0.0699897892 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_009486_080 |
0.0702450183 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 6 |
Hb_001001_150 |
0.0727272254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000086_150 |
0.0764572554 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007894_160 |
0.0785455285 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 9 |
Hb_005402_040 |
0.0799417397 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 10 |
Hb_004064_050 |
0.0803982958 |
- |
- |
Structural maintenance of chromosome, putative [Ricinus communis] |
| 11 |
Hb_002350_010 |
0.0826707583 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 12 |
Hb_003436_020 |
0.0828866493 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001789_040 |
0.0853710012 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 14 |
Hb_027472_220 |
0.0854656026 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_001454_170 |
0.0868909764 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 16 |
Hb_000473_110 |
0.0870254683 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 17 |
Hb_000671_070 |
0.0873147025 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 18 |
Hb_001318_070 |
0.0875786639 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000243_130 |
0.0876099748 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 20 |
Hb_000035_170 |
0.0881399856 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |