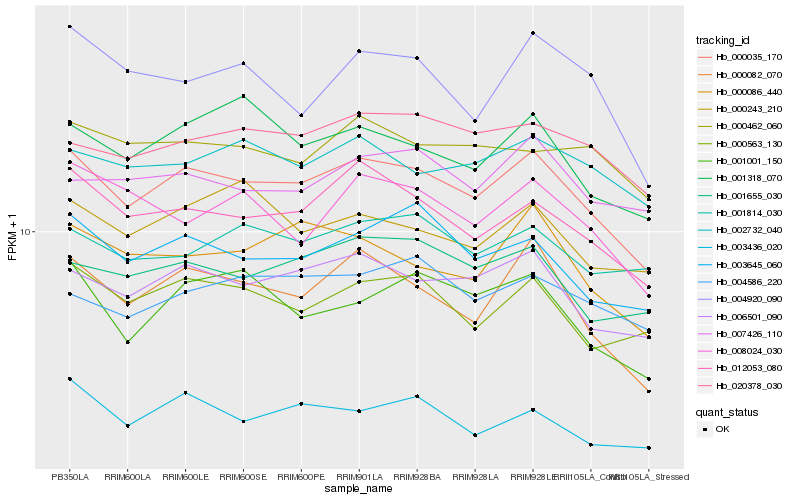
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_170 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 2 |
Hb_006501_090 |
0.0532357333 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 3 |
Hb_012053_080 |
0.0570057127 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 4 |
Hb_000082_070 |
0.0627768302 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 5 |
Hb_003645_060 |
0.0658903422 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000563_130 |
0.0694643581 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 7 |
Hb_001655_030 |
0.0702801281 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000086_440 |
0.0707624739 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 9 |
Hb_001318_070 |
0.0713213199 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_020378_030 |
0.0729262284 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 11 |
Hb_001001_150 |
0.0755112772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002732_040 |
0.0763278519 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004920_090 |
0.077647386 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_008024_030 |
0.0778656341 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 15 |
Hb_003436_020 |
0.0785579664 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000243_210 |
0.0786299385 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_004586_220 |
0.079602914 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 18 |
Hb_007426_110 |
0.0796325108 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_001814_030 |
0.079794551 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 20 |
Hb_000462_060 |
0.080291637 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |