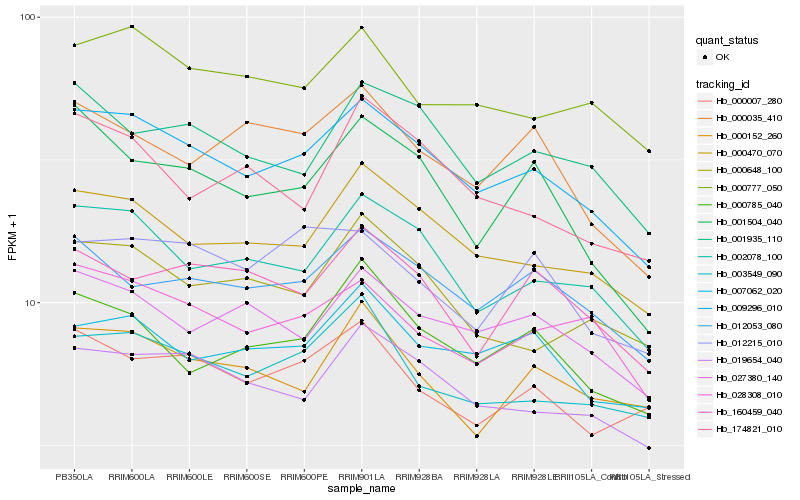
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009296_010 |
0.0 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 2 |
Hb_000470_070 |
0.0514476126 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 3 |
Hb_002078_100 |
0.0529674484 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 4 |
Hb_019654_040 |
0.0565215601 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 5 |
Hb_027380_140 |
0.0669169875 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 6 |
Hb_012053_080 |
0.0672389273 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 7 |
Hb_001935_110 |
0.0689545384 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 8 |
Hb_028308_010 |
0.0694767616 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007062_020 |
0.069872246 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 10 |
Hb_000777_050 |
0.0713084503 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 11 |
Hb_003549_090 |
0.0716688799 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 12 |
Hb_000785_040 |
0.0734544659 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 13 |
Hb_001504_040 |
0.0752464308 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 14 |
Hb_000648_100 |
0.0782943289 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 15 |
Hb_160459_040 |
0.0784554967 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 16 |
Hb_000007_280 |
0.0797175777 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 17 |
Hb_000035_410 |
0.0803738317 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 18 |
Hb_174821_010 |
0.0819913907 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 19 |
Hb_000152_260 |
0.0820999917 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 20 |
Hb_012215_010 |
0.0825999071 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |