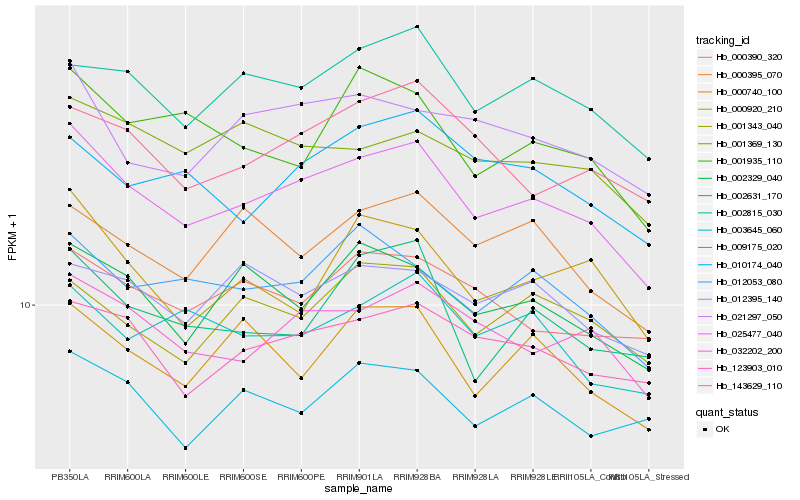
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025477_040 |
0.0 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 2 |
Hb_032202_200 |
0.0607566328 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_123903_010 |
0.0664625701 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002815_030 |
0.0680545136 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 5 |
Hb_012053_080 |
0.0695994522 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 6 |
Hb_002329_040 |
0.0704336099 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 7 |
Hb_021297_050 |
0.0734040857 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 8 |
Hb_143629_110 |
0.0741814285 |
- |
- |
glutamyl-tRNA synthetase, cytoplasmic, putative [Ricinus communis] |
| 9 |
Hb_000390_320 |
0.075121788 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-1b-like [Jatropha curcas] |
| 10 |
Hb_001343_040 |
0.0777612405 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 11 |
Hb_000740_100 |
0.07820664 |
- |
- |
calpain, putative [Ricinus communis] |
| 12 |
Hb_000395_070 |
0.078541167 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003645_060 |
0.0790184125 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001935_110 |
0.0792137251 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 15 |
Hb_000920_210 |
0.0794574202 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 16 |
Hb_010174_040 |
0.0795471665 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 17 |
Hb_012395_140 |
0.0797653839 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 18 |
Hb_009175_020 |
0.0798255581 |
- |
- |
PREDICTED: uncharacterized protein LOC101303140 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_002631_170 |
0.0804441374 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 20 |
Hb_001369_130 |
0.0804712544 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |