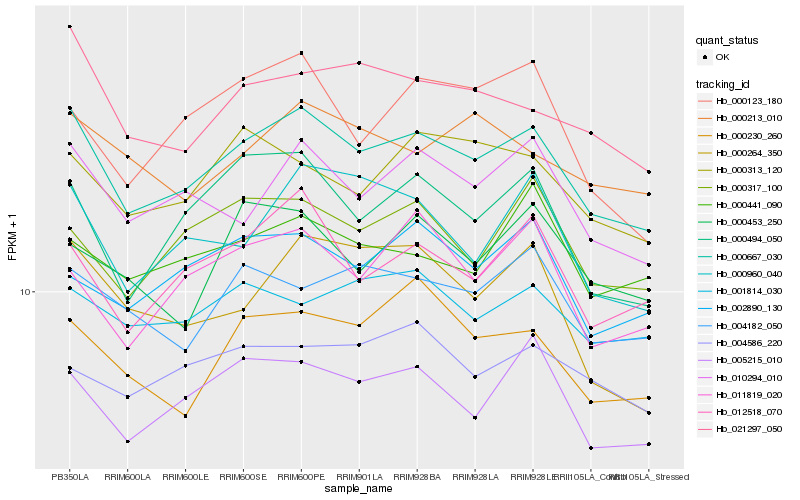
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000667_030 |
0.0 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 2 |
Hb_010294_010 |
0.0634225612 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 3 |
Hb_005215_010 |
0.0652132959 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_012518_070 |
0.0665322061 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 5 |
Hb_021297_050 |
0.0675399276 |
- |
- |
PREDICTED: signal recognition particle receptor subunit alpha [Jatropha curcas] |
| 6 |
Hb_000494_050 |
0.0699653099 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_000317_100 |
0.073764613 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 8 |
Hb_000960_040 |
0.0742433137 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 9 |
Hb_004182_050 |
0.0761885245 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000313_120 |
0.0764275082 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 11 |
Hb_000230_260 |
0.0776032172 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 12 |
Hb_002890_130 |
0.0788576851 |
- |
- |
tip120, putative [Ricinus communis] |
| 13 |
Hb_001814_030 |
0.078899313 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 14 |
Hb_011819_020 |
0.0798180497 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000213_010 |
0.0813102687 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 16 |
Hb_000441_090 |
0.0819940057 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 17 |
Hb_004586_220 |
0.0821360835 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000123_180 |
0.0829627437 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000453_250 |
0.083430019 |
- |
- |
PREDICTED: uncharacterized protein LOC105637185 [Jatropha curcas] |
| 20 |
Hb_000264_350 |
0.0834896715 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |