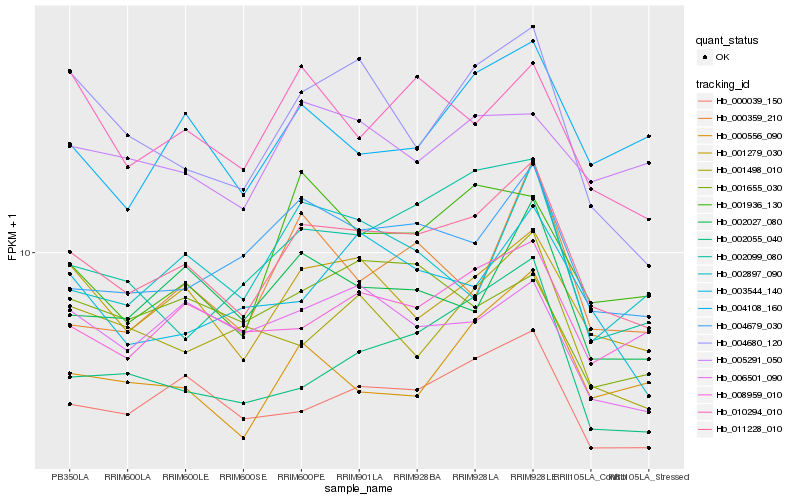
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011228_010 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001279_030 |
0.0571375029 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 3 |
Hb_000039_150 |
0.0614168986 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004679_030 |
0.0659437147 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 5 |
Hb_002027_080 |
0.0687243158 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 6 |
Hb_008959_010 |
0.0690418282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002055_040 |
0.0705367808 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 8 |
Hb_001936_130 |
0.0749224957 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 9 |
Hb_000556_090 |
0.0771547026 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010294_010 |
0.0774750777 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 11 |
Hb_003544_140 |
0.077651288 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_004108_160 |
0.0786491791 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 13 |
Hb_000359_210 |
0.0799188882 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006501_090 |
0.0799678687 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 15 |
Hb_002099_080 |
0.0800849315 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 16 |
Hb_004680_120 |
0.0809260507 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 17 |
Hb_005291_050 |
0.0819942652 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002897_090 |
0.0832504477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001498_010 |
0.0836200763 |
- |
- |
PREDICTED: uncharacterized protein LOC105648994 [Jatropha curcas] |
| 20 |
Hb_001655_030 |
0.0857942847 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |