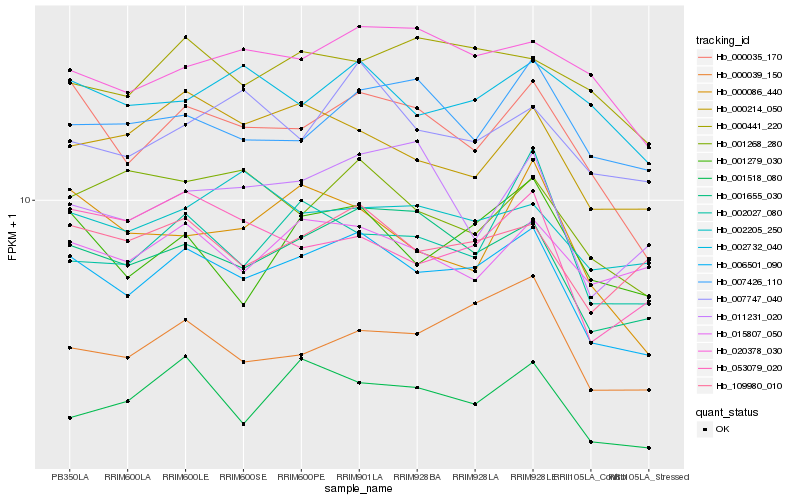
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006501_090 |
0.0 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 2 |
Hb_001655_030 |
0.0468593786 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000035_170 |
0.0532357333 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 4 |
Hb_109980_010 |
0.0647448631 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001518_080 |
0.0662131423 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 6 |
Hb_001279_030 |
0.0663180367 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 7 |
Hb_000039_150 |
0.0668150616 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000214_050 |
0.0678695123 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_020378_030 |
0.0718226906 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 10 |
Hb_053079_020 |
0.0721881885 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 11 |
Hb_015807_050 |
0.0723290711 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 12 |
Hb_007747_040 |
0.073304019 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 13 |
Hb_000441_220 |
0.073401914 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_002732_040 |
0.0739370664 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007426_110 |
0.0740526902 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_000086_440 |
0.0742553591 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 17 |
Hb_002027_080 |
0.0742691634 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 18 |
Hb_001268_280 |
0.0746529175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002205_250 |
0.0755917467 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011231_020 |
0.0759644721 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |