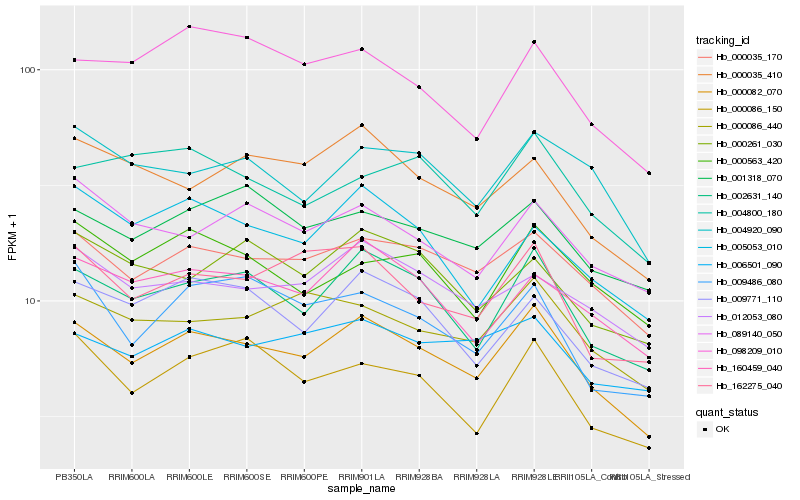
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000082_070 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 2 |
Hb_002631_140 |
0.0557903392 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000035_170 |
0.0627768302 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 4 |
Hb_160459_040 |
0.0790419902 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 5 |
Hb_162275_040 |
0.0796093579 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000086_440 |
0.081263681 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 7 |
Hb_009771_110 |
0.0822910533 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 8 |
Hb_012053_080 |
0.084311431 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 9 |
Hb_006501_090 |
0.0864186179 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 10 |
Hb_001318_070 |
0.0867997114 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004920_090 |
0.0874988388 |
- |
- |
PREDICTED: probable glutamyl endopeptidase, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000035_410 |
0.0880197978 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 13 |
Hb_089140_050 |
0.0892767247 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 14 |
Hb_000086_150 |
0.08934233 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_098209_010 |
0.0914568012 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 16 |
Hb_009486_080 |
0.0917734041 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 17 |
Hb_000563_420 |
0.092060001 |
- |
- |
PREDICTED: acyl-CoA dehydrogenase family member 10 [Jatropha curcas] |
| 18 |
Hb_004800_180 |
0.092363384 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 19 |
Hb_000261_030 |
0.0931009397 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 20 |
Hb_005053_010 |
0.0932504605 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |