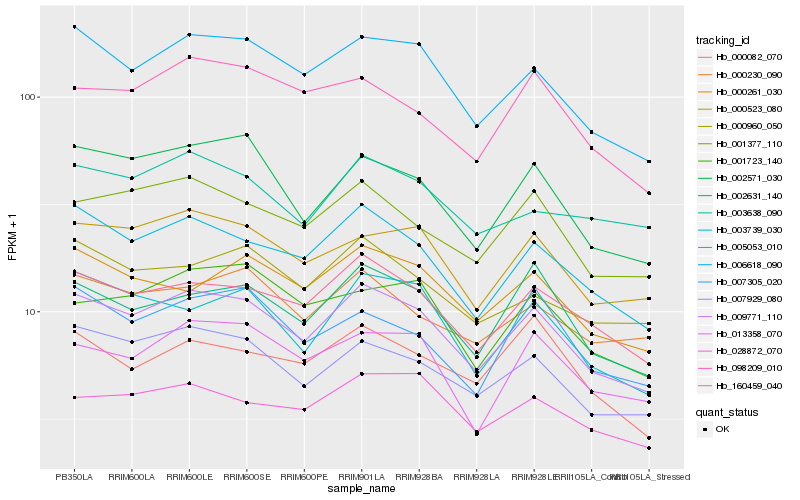
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009771_110 |
0.0 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 2 |
Hb_006618_090 |
0.0459966959 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 3 |
Hb_005053_010 |
0.0480623557 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 4 |
Hb_160459_040 |
0.0521042291 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 5 |
Hb_007929_080 |
0.0544382314 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 6 |
Hb_002571_030 |
0.0609865984 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 7 |
Hb_000523_080 |
0.0614907625 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 8 |
Hb_002631_140 |
0.0631560839 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001377_110 |
0.0631729577 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 10 |
Hb_003739_030 |
0.0705961074 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 11 |
Hb_000261_030 |
0.071937603 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 12 |
Hb_013358_070 |
0.0733492616 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_028872_070 |
0.0740814678 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_098209_010 |
0.0752046891 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 15 |
Hb_000960_050 |
0.0759305838 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 16 |
Hb_000230_090 |
0.0759767152 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001723_140 |
0.0777516636 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_003638_090 |
0.0808560951 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 19 |
Hb_007305_020 |
0.0813980785 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 20 |
Hb_000082_070 |
0.0822910533 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |