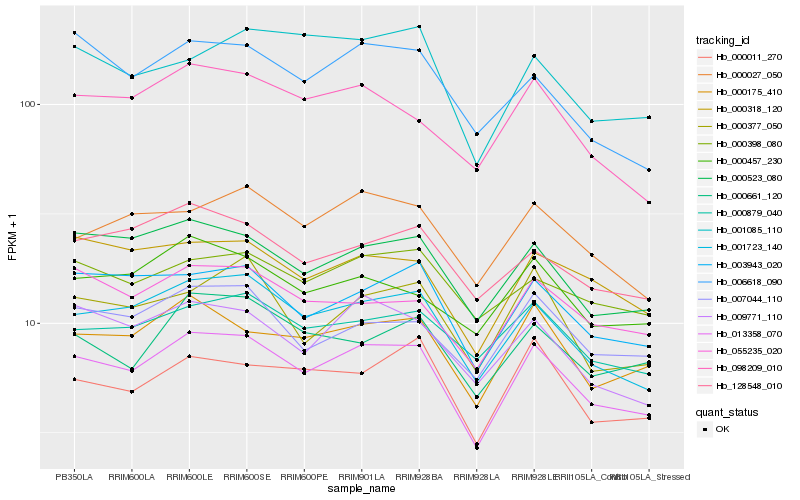
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013358_070 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_001723_140 |
0.0521294957 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_000523_080 |
0.0653833793 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 4 |
Hb_000175_410 |
0.0662859488 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_003943_020 |
0.0675966655 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009771_110 |
0.0733492616 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 7 |
Hb_000318_120 |
0.0738424376 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007044_110 |
0.075436544 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000027_050 |
0.0755279524 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 10 |
Hb_055235_020 |
0.0765392706 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 11 |
Hb_098209_010 |
0.0783380775 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 12 |
Hb_000879_040 |
0.0795175479 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 13 |
Hb_000011_270 |
0.0798008882 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 14 |
Hb_000377_050 |
0.0804533601 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 15 |
Hb_128548_010 |
0.0815757887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000661_120 |
0.0820730023 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 17 |
Hb_000457_230 |
0.0821315318 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006618_090 |
0.0828442052 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 19 |
Hb_000398_080 |
0.0830073211 |
- |
- |
tip120, putative [Ricinus communis] |
| 20 |
Hb_001085_110 |
0.083118041 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |