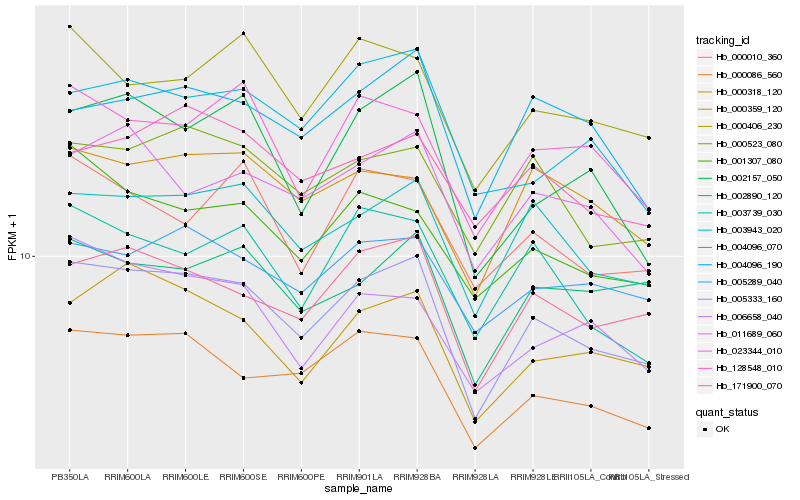
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005333_160 |
0.0 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 2 |
Hb_171900_070 |
0.066251096 |
- |
- |
- |
| 3 |
Hb_003943_020 |
0.0675597704 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000086_560 |
0.0688512993 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000359_120 |
0.0768789679 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000523_080 |
0.0801270076 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 7 |
Hb_011689_060 |
0.0854344728 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 8 |
Hb_005289_040 |
0.0878592935 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 9 |
Hb_000318_120 |
0.0880655108 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002157_050 |
0.0882361298 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 11 |
Hb_000406_230 |
0.0888396015 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 12 |
Hb_001307_080 |
0.0892808104 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 13 |
Hb_003739_030 |
0.08959613 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 14 |
Hb_023344_010 |
0.0901157195 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004096_190 |
0.092081445 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 16 |
Hb_004096_070 |
0.0933434734 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_002890_120 |
0.0936454364 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 18 |
Hb_000010_360 |
0.0946686546 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 19 |
Hb_128548_010 |
0.0948703067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006658_040 |
0.0951080841 |
- |
- |
hypothetical protein POPTR_0006s19640g [Populus trichocarpa] |