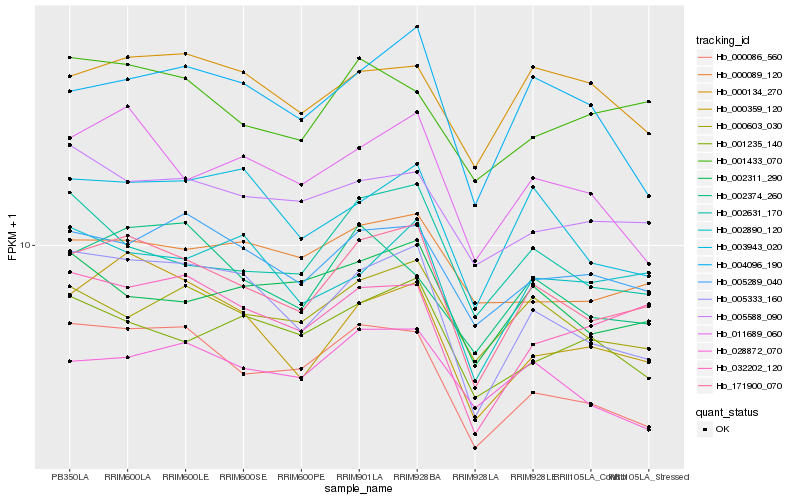
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171900_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_005333_160 |
0.066251096 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 3 |
Hb_000359_120 |
0.0802759851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005289_040 |
0.0813651686 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 5 |
Hb_000086_560 |
0.0816559933 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_032202_120 |
0.0843556628 |
- |
- |
PREDICTED: uncharacterized protein LOC105643059 isoform X2 [Jatropha curcas] |
| 7 |
Hb_011689_060 |
0.0848346932 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 8 |
Hb_000603_030 |
0.0881202297 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 9 |
Hb_003943_020 |
0.0902699041 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002631_170 |
0.0911819271 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 11 |
Hb_001433_070 |
0.0916170403 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 12 |
Hb_002374_260 |
0.0918117115 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 13 |
Hb_028872_070 |
0.0922175768 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004096_190 |
0.0924893028 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 15 |
Hb_002311_290 |
0.0938951232 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 16 |
Hb_002890_120 |
0.0941792652 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 17 |
Hb_000089_120 |
0.0942451824 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_005588_090 |
0.0950310823 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 19 |
Hb_001235_140 |
0.0963372815 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 20 |
Hb_000134_270 |
0.097140769 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |