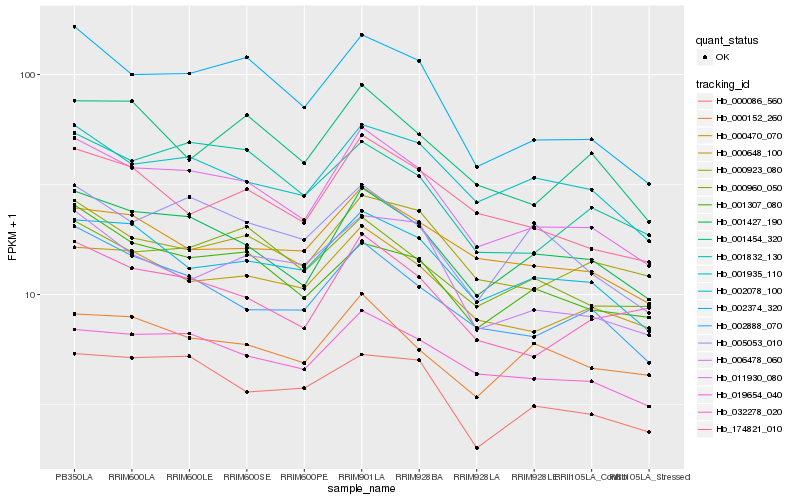
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011930_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 2 |
Hb_001427_190 |
0.051266689 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 3 |
Hb_002374_320 |
0.0555378609 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 4 |
Hb_019654_040 |
0.0630143569 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000648_100 |
0.0634531605 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 6 |
Hb_002078_100 |
0.0676698755 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 7 |
Hb_002888_070 |
0.0701538778 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 8 |
Hb_001935_110 |
0.0710423412 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 9 |
Hb_174821_010 |
0.0726040771 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 10 |
Hb_032278_020 |
0.077001304 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000470_070 |
0.0774629353 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 12 |
Hb_001307_080 |
0.0775534619 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 13 |
Hb_000923_080 |
0.0776442863 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 14 |
Hb_000960_050 |
0.0794884542 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 15 |
Hb_006478_060 |
0.0800053267 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005053_010 |
0.0812726928 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 17 |
Hb_000086_560 |
0.0813251795 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001832_130 |
0.082600558 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 19 |
Hb_001454_320 |
0.0839346407 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000152_260 |
0.0845258297 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |